R 数据类型
向量
概念类似于Python里的列表,只能存储相同类型 的数据。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 x = 1 x <- c ( 1 , 2 , 3 ) mode( x) [ 1 ] "numeric" z <- c ( T , F , T ) mode( z) [ 1 ] "logical" x <- c ( 1 : 100 ) seq( from= 1 , to= 20 , by= 3 ) [ 1 ] 1 4 7 10 13 16 19 ? seq rep ( x, 2 ) [ 1 ] 1 2 3 1 2 3 rep ( x, c ( 1 , 2 , 3 ) ) [ 1 ] 1 2 2 3 3 3 y <- c ( 2 , 3 , 4 ) x+ y [ 1 ] 3 5 7 x** 2 [ 1 ] 1 4 9
向量索引
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 x <- c ( 1 : 100 ) length ( x) [ 1 ] 100 x[ 1 ] [ 1 ] 1 x[ 0 ] integer( 0 ) x[ - 10 ] [ 1 ] 1 2 3 4 5 6 7 8 9 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 [ 25 ] 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 [ 49 ] 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 [ 73 ] 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 [ 97 ] 98 99 100 x[ - c ( 10 , 1 ) ] [ 1 ] 2 3 4 5 6 7 8 9 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 [ 25 ] 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 [ 49 ] 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 [ 73 ] 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 [ 97 ] 99 100 x[ c ( 2 : 10 ) ] [ 1 ] 2 3 4 5 6 7 8 9 10 x[ c ( 1 , 2 , 5 , 10 ) ] [ 1 ] 1 2 5 10 x[ c ( 1 , 2 , 5 , 10 , 10 , 10 ) ] [ 1 ] 1 2 5 10 10 10 x[ - 10 , - 1 ] Error in x[ - 10 , - 1 ] : 量度数目不对 x[ x> 3 ] [ 1 ] 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 [ 25 ] 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 [ 49 ] 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 [ 73 ] 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 [ 97 ] 100 x[ x> 5 & x< 8 ] [ 1 ] 6 7 y <- c ( 1 : 10 ) y[ c ( T ) ] [ 1 ] 1 2 3 4 5 6 7 8 9 10 y[ c ( F ) ] integer( 0 ) y[ c ( T , F ) ] [ 1 ] 1 3 5 7 9 y[ c ( T , T ) ] [ 1 ] 1 2 3 4 5 6 7 8 9 10 y[ c ( F , T ) ] [ 1 ] 2 4 6 8 10 y[ c ( T , T , T , T , T , T , T , T , T , T , T , T ) ] [ 1 ] 1 2 3 4 5 6 7 8 9 10 NA NA z <- c ( 'a' , 'b' , 'c' , 'd' , 'e' ) z [ 1 ] "a" "b" "c" "d" "e" names ( z) <- c ( 'one' , 'two' , 'three' , 'four' , 'five' ) z one two three four five "a" "b" "c" "d" "e" z[ 'one' ] one "a" z[ 'one' ] <- 'b' z one two three four five "b" "b" "c" "d" "e" z[ c ( 2 , 3 ) ] <- c ( 'x' , 'y' ) z one two three four five "b" "x" "y" "d" "e" z[ c ( 6 , 7 ) ] <- c ( 'x' , 'y' ) z one two three four five "b" "x" "y" "d" "e" "x" "y" 'b' %in% z [ 1 ] TRUE z[ z %in% c ( 'a' , 'b' , 'd' , 'e' ) ] one four five "b" "d" "e" append( x = z, values = 'zzz' , after = 0 ) one two three four five "zzz" "b" "x" "y" "d" "e" "x" "y" append( x = z, values = 'zzz' , after = 1 ) one two three four five "b" "zzz" "x" "y" "d" "e" "x" "y" append( x = z, values = 'zzz' , after = 2 ) one two three four five "b" "x" "zzz" "y" "d" "e" "x" "y" append( x = z, values = 'zzz' , after = 5 ) one two three four five "b" "x" "y" "d" "e" "zzz" "x" "y" append( x = z, values = 'zzz' , after = 10 ) one two three four five "b" "x" "y" "d" "e" "x" "y" "zzz" rm( z) z 错误: 找不到对象'z' y 5 "1" "2" "3" "4" "5" "6" "7" "8" "9" "e" y[ - c ( 10 ) ] "1" "2" "3" "4" "5" "6" "7" "8" "9" y <- y[ - c ( 10 ) ] y "1" "2" "3" "4" "5" "6" "7" "8" "9" euro ATS BEF DEM ESP FIM FRF IEP ITL 13.760300 40.339900 1.955830 166.386000 5.945730 6.559570 0.787564 1936.270000 LUF NLG PTE 40.339900 2.203710 200.482000 euro[ 'PTE' ] PTE 200.482
矩阵与数组
矩阵是二维 的,有行和列,每个元素类型要相同,可以看成向量的扩充。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 head( iris3) , , Setosa Sepal L. Sepal W. Petal L. Petal W. [ 1 , ] 5.1 3.5 1.4 0.2 [ 2 , ] 4.9 3.0 1.4 0.2 [ 3 , ] 4.7 3.2 1.3 0.2 [ 4 , ] 4.6 3.1 1.5 0.2 [ 5 , ] 5.0 3.6 1.4 0.2 [ 6 , ] 5.4 3.9 1.7 0.4 , , Versicolor Sepal L. Sepal W. Petal L. Petal W. [ 1 , ] 7.0 3.2 4.7 1.4 [ 2 , ] 6.4 3.2 4.5 1.5 [ 3 , ] 6.9 3.1 4.9 1.5 [ 4 , ] 5.5 2.3 4.0 1.3 [ 5 , ] 6.5 2.8 4.6 1.5 [ 6 , ] 5.7 2.8 4.5 1.3 , , Virginica Sepal L. Sepal W. Petal L. Petal W. [ 1 , ] 6.3 3.3 6.0 2.5 [ 2 , ] 5.8 2.7 5.1 1.9 [ 3 , ] 7.1 3.0 5.9 2.1 [ 4 , ] 6.3 2.9 5.6 1.8 [ 5 , ] 6.5 3.0 5.8 2.2 [ 6 , ] 7.6 3.0 6.6 2.1 head( state.x77) Population Income Illiteracy Life Exp Murder HS Grad Frost Area Alabama 3615 3624 2.1 69.05 15.1 41.3 20 50708 Alaska 365 6315 1.5 69.31 11.3 66.7 152 566432 Arizona 2212 4530 1.8 70.55 7.8 58.1 15 113417 Arkansas 2110 3378 1.9 70.66 10.1 39.9 65 51945 California 21198 5114 1.1 71.71 10.3 62.6 20 156361 Colorado 2541 4884 0.7 72.06 6.8 63.9 166 103766 heatmap( state.x77) m <- matrix( 1 : 20 , 4 , 5 ) m [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 1 5 9 13 17 [ 2 , ] 2 6 10 14 18 [ 3 , ] 3 7 11 15 19 [ 4 , ] 4 8 12 16 20 m <- matrix( 1 : 20 , 4 , 5 , byrow = T ) m [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 1 2 3 4 5 [ 2 , ] 6 7 8 9 10 [ 3 , ] 11 12 13 14 15 [ 4 , ] 16 17 18 19 20 m <- matrix( c ( 1 : 20 ) , 4 , 6 , byrow = T ) Warning message: In matrix( c ( 1 : 20 ) , 4 , 6 , byrow = T ) : 数据长度[ 20 ] 不是矩阵列数[ 6 ] 的整倍数 m <- matrix( c ( 1 : 20 ) , 3 , 5 , byrow = T ) Warning message: In matrix( c ( 1 : 20 ) , 3 , 5 , byrow = T ) : 数据长度[ 20 ] 不是矩阵行数[ 3 ] 的整倍 m <- matrix( c ( 1 : 20 ) , 4 , 4 , byrow = T ) m [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ 1 , ] 1 2 3 4 [ 2 , ] 5 6 7 8 [ 3 , ] 9 10 11 12 [ 4 , ] 13 14 15 16 m <- matrix( c ( 1 : 20 ) , 4 , 10 , byrow = T ) m [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ , 6 ] [ , 7 ] [ , 8 ] [ , 9 ] [ , 10 ] [ 1 , ] 1 2 3 4 5 6 7 8 9 10 [ 2 , ] 11 12 13 14 15 16 17 18 19 20 [ 3 , ] 1 2 3 4 5 6 7 8 9 10 [ 4 , ] 11 12 13 14 15 16 17 18 19 20 m <- matrix( 1 : 20 , 2 , 4 ) m [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ 1 , ] 1 3 5 7 [ 2 , ] 2 4 6 8 cnames <- c ( 'C1' , 'C2' , 'C3' , 'C4' ) rnames <- c ( 'R1' , 'R2' ) dimnames ( m) <- list ( rnames, cnames) m C1 C2 C3 C4 R1 1 3 5 7 R2 2 4 6 8 dim ( m) [ 1 ] 2 4 dim ( m) <- c ( 1 , 8 ) m [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ , 6 ] [ , 7 ] [ , 8 ] [ 1 , ] 1 2 3 4 5 6 7 8 dim ( m) <- c ( 4 , 2 ) m [ , 1 ] [ , 2 ] [ 1 , ] 1 5 [ 2 , ] 2 6 [ 3 , ] 3 7 [ 4 , ] 4 8 x <- 1: 9 dim ( x) <- c ( 3 , 3 ) x [ , 1 ] [ , 2 ] [ , 3 ] [ 1 , ] 1 4 7 [ 2 , ] 2 5 8 [ 3 , ] 3 6 9
矩阵索引
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 x [ , 1 ] [ , 2 ] [ , 3 ] [ 1 , ] 1 4 7 [ 2 , ] 2 5 8 [ 3 , ] 3 6 9 x[ 1 , 1 ] [ 1 ] 1 x[ 1 , 2 ] [ 1 ] 4 x[ 1 , ] [ 1 ] 1 4 7 x[ , 1 ] [ 1 ] 1 2 3 x[ 1 , c ( 2 , 3 ) ] [ 1 ] 4 7 x[ c ( 1 , 2 ) , c ( 1 , 2 ) ] [ , 1 ] [ , 2 ] [ 1 , ] 1 4 [ 2 , ] 2 5 x[ 1 ] [ 1 ] 1 x[ 2 ] [ 1 ] 2 x[ - 1 ] [ 1 ] 2 3 4 5 6 7 8 9 x[ - 1 , ] [ , 1 ] [ , 2 ] [ , 3 ] [ 1 , ] 2 5 8 [ 2 , ] 3 6 9 x[ - 1 , 2 ] [ 1 ] 5 6 x[ 1 , - 2 ] [ 1 ] 1 7 head( state.x77) Population Income Illiteracy Life Exp Murder HS Grad Frost Area Alabama 3615 3624 2.1 69.05 15.1 41.3 20 50708 Alaska 365 6315 1.5 69.31 11.3 66.7 152 566432 Arizona 2212 4530 1.8 70.55 7.8 58.1 15 113417 Arkansas 2110 3378 1.9 70.66 10.1 39.9 65 51945 California 21198 5114 1.1 71.71 10.3 62.6 20 156361 Colorado 2541 4884 0.7 72.06 6.8 63.9 166 103766 state.x77[ 'California' , ] Population Income Illiteracy Life Exp Murder HS Grad Frost Area 21198.00 5114.00 1.10 71.71 10.30 62.60 20.00 156361.00
矩阵运算与线性代数中一致。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 m [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 1 5 9 13 17 [ 2 , ] 2 6 10 14 18 [ 3 , ] 3 7 11 15 19 [ 4 , ] 4 8 12 16 20 n [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 2 6 10 14 18 [ 2 , ] 3 7 11 15 19 [ 3 , ] 4 8 12 16 20 [ 4 , ] 5 9 13 17 21 m+ n [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 3 11 19 27 35 [ 2 , ] 5 13 21 29 37 [ 3 , ] 7 15 23 31 39 [ 4 , ] 9 17 25 33 41 m* n [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 2 30 90 182 306 [ 2 , ] 6 42 110 210 342 [ 3 , ] 12 56 132 240 380 [ 4 , ] 20 72 156 272 420 m %*% t( n) [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ 1 , ] 610 655 700 745 [ 2 , ] 660 710 760 810 [ 3 , ] 710 765 820 875 [ 4 , ] 760 820 880 940 rowSums( m) [ 1 ] 45 50 55 60 colSums( m) [ 1 ] 10 26 42 58 74 rowMeans( m) [ 1 ] 9 10 11 12 colMeans( m) [ 1 ] 2.5 6.5 10.5 14.5 18.5 n [ , 1 ] [ , 2 ] [ , 3 ] [ 1 , ] 1 4 7 [ 2 , ] 2 5 8 [ 3 , ] 3 6 9 diag( n) [ 1 ] 1 5 9
数组即多维矩阵。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 dim1 <- c ( 'A1' , 'A2' , 'A3' , 'A4' , 'A5' ) dim2 <- c ( 'B1' , 'B2' ) dim3 <- c ( 'C1' , 'C2' ) x <- array( 1 : 20 , c ( 5 , 2 , 2 ) , dimnames = list ( dim1, dim2, dim3) ) x , , C1 B1 B2 A1 1 6 A2 2 7 A3 3 8 A4 4 9 A5 5 10 , , C2 B1 B2 A1 11 16 A2 12 17 A3 13 18 A4 14 19 A5 15 20
列表
R中最复杂的数据结构,可以用来存储不同的数据,例如向量、矩阵、数据框、列表等。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 a <- 1: 20 b <- c ( 'a' , 'b' , 'c' , 'd' ) mode( mtcars) [ 1 ] "list" c <- mtcarsd <- matrix( 1 : 20 , 4 , 5 ) mlist <- list ( a, b, c , d) mlist [[ 1 ] ] [ 1 ] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 [[ 2 ] ] [ 1 ] "a" "b" "c" "d" [[ 3 ] ] mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 ... [[ 4 ] ] [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 1 5 9 13 17 [ 2 , ] 2 6 10 14 18 [ 3 , ] 3 7 11 15 19 [ 4 , ] 4 8 12 16 20 mlist <- list ( first= a, second= b, third= c , fourth= d) mlist $ first [ 1 ] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 $ second[ 1 ] "a" "b" "c" "d" $ third mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 ... $ fourth [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ 1 , ] 1 5 9 13 17 [ 2 , ] 2 6 10 14 18 [ 3 , ] 3 7 11 15 19 [ 4 , ] 4 8 12 16 20 mlist <- list ( 'first' = a) mlist $ first [ 1 ] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
列表索引
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 mlist[ 'second' ] $ second[ 1 ] "a" "b" "c" "d" mlist[ 2 ] $ second[ 1 ] "a" "b" "c" "d" class ( mlist[ 2 ] ) [ 1 ] "list" mlist[[ 2 ] ] [ 1 ] "a" "b" "c" "d" class ( mlist[[ 2 ] ] ) [ 1 ] "character" class ( mlist$ third) [ 1 ] "data.frame" class ( mlist$ fourth) [ 1 ] "matrix" "array" mlist[ 5 ] <- iris Warning message: In mlist[ 5 ] <- iris : number of items to replace is not a multiple of replacement length mlist[[ 5 ] ] <- iris mlist <- mlist[ - 5 ] mlist[ 4 ] <- NULL mlist $ first [ 1 ] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 $ second[ 1 ] "a" "b" "c" "d" $ third mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 ...
数据框
类似于表格,每一列长度和数据类型必须相同,且必须命名,每行可不相同。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 class ( iris) [ 1 ] "data.frame" class ( mtcars) [ 1 ] "data.frame" iris Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa... mtcars mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 ... state <- data.frame( state.name, state.abb, state.x77) state state.name state.abb Population Income Illiteracy Life.Exp Murder HS.Grad Frost Alabama Alabama AL 3615 3624 2.1 69.05 15.1 41.3 20 Alaska Alaska AK 365 6315 1.5 69.31 11.3 66.7 152 Arizona Arizona AZ 2212 4530 1.8 70.55 7.8 58.1 15 ... rownames( state) [ 1 ] "Alabama" "Alaska" "Arizona" "Arkansas" "California" [ 6 ] "Colorado" "Connecticut" "Delaware" "Florida" "Georgia" [ 11 ] "Hawaii" "Idaho" "Illinois" "Indiana" "Iowa" [ 16 ] "Kansas" "Kentucky" "Louisiana" "Maine" "Maryland" [ 21 ] "Massachusetts" "Michigan" "Minnesota" "Mississippi" "Missouri" [ 26 ] "Montana" "Nebraska" "Nevada" "New Hampshire" "New Jersey" [ 31 ] "New Mexico" "New York" "North Carolina" "North Dakota" "Ohio" [ 36 ] "Oklahoma" "Oregon" "Pennsylvania" "Rhode Island" "South Carolina" [ 41 ] "South Dakota" "Tennessee" "Texas" "Utah" "Vermont" [ 46 ] "Virginia" "Washington" "West Virginia" "Wisconsin" "Wyoming" colnames( state) [ 1 ] "state.name" "state.abb" "Population" "Income" "Illiteracy" "Life.Exp" "Murder" [ 8 ] "HS.Grad" "Frost" "Area" state[ 1 ] state.name Alabama Alabama Alaska Alaska Arizona Arizona ... state[ 'state.abb' ] state.abb Alabama AL Alaska AK Arizona AZ ... state$ Murder [ 1 ] 15.1 11.3 7.8 10.1 10.3 6.8 3.1 6.2 10.7 13.9 6.2 5.3 10.3 7.1 2.3 4.5 10.6 13.2 2.7 [ 20 ] 8.5 3.3 11.1 2.3 12.5 9.3 5.0 2.9 11.5 3.3 5.2 9.7 10.9 11.1 1.4 7.4 6.4 4.2 6.1 [ 39 ] 2.4 11.6 1.7 11.0 12.2 4.5 5.5 9.5 4.3 6.7 3.0 6.9 state[ , 'state.abb' ] [ 1 ] "AL" "AK" "AZ" "AR" "CA" "CO" "CT" "DE" "FL" "GA" "HI" "ID" "IL" "IN" "IA" "KS" "KY" "LA" "ME" [ 20 ] "MD" "MA" "MI" "MN" "MS" "MO" "MT" "NE" "NV" "NH" "NJ" "NM" "NY" "NC" "ND" "OH" "OK" "OR" "PA" [ 39 ] "RI" "SC" "SD" "TN" "TX" "UT" "VT" "VA" "WA" "WV" "WI" "WY" state[ 'California' , ] state.name state.abb Population Income Illiteracy Life.Exp Murder HS.Grad Frost Area California California CA 21198 5114 1.1 71.71 10.3 62.6 20 156361 women height weight 1 58 115 2 59 117 3 60 120 ... class ( state[ 'state.abb' ] ) [ 1 ] "data.frame" class ( state[[ 'state.abb' ] ] ) [ 1 ] "character" state[[ 'state.abb' ] ] [ 1 ] "AL" "AK" "AZ" "AR" "CA" "CO" "CT" "DE" "FL" "GA" "HI" "ID" "IL" "IN" "IA" "KS" "KY" "LA" "ME" [ 20 ] "MD" "MA" "MI" "MN" "MS" "MO" "MT" "NE" "NV" "NH" "NJ" "NM" "NY" "NC" "ND" "OH" "OK" "OR" "PA" [ 39 ] "RI" "SC" "SD" "TN" "TX" "UT" "VT" "VA" "WA" "WV" "WI" "WY" state[ 'state.abb' ] state.abb Alabama AL Alaska AK Arizona AZ ...
因子
因子是R中重要的概念。
R中变量类型
名义型变量:没有顺序分别,相互独立,如省份名。
有序型变量:介于二者之间,不同值之间有顺序关系,但不是连续的数量变化,如good、better、best。
连续型变量:某个范围中的任意值,如存储年龄、身高、GDP的变量。
一般,字符型数据多为名义型变量,数值型数据多为连续型变量。
名义型变量和有序型变量在R中称为因子,这些变量的可能值称为一个水平level,如good,better,best都称为一个level。
因子的最大作用是用来分类,计算频数和频率。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 state.region [ 1 ] South West West South West West [ 7 ] Northeast South South South West West [ 13 ] North Central North Central North Central North Central South South ... Levels: Northeast South North Central West state.division [ 1 ] East South Central Pacific Mountain West South Central [ 5 ] Pacific Mountain New England South Atlantic [ 9 ] South Atlantic South Atlantic Pacific Mountain ... 9 Levels: New England Middle Atlantic South Atlantic ... Pacificf <- factor( c ( 1 , 1 , 2 , 2 , 2 , 3 , 4 ) ) f [ 1 ] 1 1 2 2 2 3 4 Levels: 1 2 3 4 mtcars$ cyl [ 1 ] 6 6 4 6 8 6 8 4 4 6 6 8 8 8 8 8 8 4 4 4 4 8 8 8 8 4 4 4 8 6 8 4 factor( mtcars$ cyl) [ 1 ] 6 6 4 6 8 6 8 4 4 6 6 8 8 8 8 8 8 4 4 4 4 8 8 8 8 4 4 4 8 6 8 4 Levels: 4 6 8 factor( c ( 'one' , 'one' , 'two' , 'three' , 'three' ) , ordered = T , levels = c ( 'one' , 'two' , 'three' , 'four' ) ) [ 1 ] one one two three threeLevels: one < two < three < four factor( c ( 'five' , 'five' , 'two' , 'three' , 'three' ) , ordered = T , levels = c ( 'one' , 'two' , 'three' , 'four' ) ) [ 1 ] < NA < NA two three three Levels: one < two < three < four plot( mtcars$ cyl) plot( factor( mtcars$ cyl) ) n <- 1: 20 cut( n, c ( seq( 0 , 20 , 5 ) ) ) [ 1 ] ( 0 , 5 ] ( 0 , 5 ] ( 0 , 5 ] ( 0 , 5 ] ( 0 , 5 ] ( 5 , 10 ] ( 5 , 10 ] ( 5 , 10 ] ( 5 , 10 ] ( 5 , 10 ] ( 10 , 15 ] [ 12 ] ( 10 , 15 ] ( 10 , 15 ] ( 10 , 15 ] ( 10 , 15 ] ( 15 , 20 ] ( 15 , 20 ] ( 15 , 20 ] ( 15 , 20 ] ( 15 , 20 ] Levels: ( 0 , 5 ] ( 5 , 10 ] ( 10 , 15 ] ( 15 , 20 ] mtcars$ wt [ 1 ] 2.620 2.875 2.320 3.215 3.440 3.460 3.570 3.190 3.150 3.440 3.440 4.070 3.730 3.780 5.250 [ 16 ] 5.424 5.345 2.200 1.615 1.835 2.465 3.520 3.435 3.840 3.845 1.935 2.140 1.513 3.170 2.770 [ 31 ] 3.570 2.780 range ( mtcars$ wt) [ 1 ] 1.513 5.424 plot( cut( mtcars$ wt, c ( seq( 1 , 6 , 1 ) ) ) )
缺失数据
NA(not available)代表缺失值,用来存储缺失数据,表示没有。注意没有不是0,可能是任何值。
NaN代表不可能值,不存在。
Inf表示无穷大,-Inf为无穷小。
在R中有相关包和函数可以处理缺失值,如VIM包。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 [ 1 ] NaN NA + 1 [ 1 ] NA 0 / 0 [ 1 ] NaN 1 / 0 [ 1 ] Inf - 1 / 0 [ 1 ] - Inf a <- c ( NA , NaN , Inf , - Inf ) is.na ( a) [ 1 ] TRUE TRUE FALSE FALSE is.infinite ( a) [ 1 ] FALSE FALSE TRUE TRUE is.nan ( a) [ 1 ] FALSE TRUE FALSE FALSE a [ 1 ] NA 1 2 3 4 5 6 7 8 9 10 sum ( a) [ 1 ] NA mean( a) [ 1 ] NA sum ( a, na.rm = T ) [ 1 ] 55 mean( a, na.rm = T ) [ 1 ] 5.5 a [ 1 ] NA 1 2 3 4 NA NA b <- na.omit( a) b [ 1 ] 1 2 3 4 attr ( , "na.action" ) [ 1 ] 1 6 7 attr ( , "class" ) [ 1 ] "omit" is.na ( b) [ 1 ] FALSE FALSE FALSE FALSE sum ( b) [ 1 ] 10 a state.name state.area 1 < NA 51609 2 < NA 589757 3 Arizona 113909 4 Arkansas 53104 5 California 158693 na.omit( a) state.name state.area 3 Arizona 113909 4 Arkansas 53104 5 California 158693
字符串
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 nchar( 'hello' ) [ 1 ] 5 a <- c ( 'hell o' , 'world' , '!' ) nchar( a) [ 1 ] 6 5 1 month.name [ 1 ] "January" "February" "March" "April" "May" "June" "July" [ 8 ] "August" "September" "October" "November" "December" nchar( month.name ) [ 1 ] 7 8 5 5 3 4 4 6 9 7 8 8 length ( month.name ) [ 1 ] 12 nchar( 12345 ) [ 1 ] 5 nchar( c ( 12 , 133 ) ) [ 1 ] 2 3 paste( 'hello' , 'world' , '!' ) [ 1 ] "hello world !" paste( 'hello' , 'world' , '!' , sep = '-' ) [ 1 ] "hello-world-!" a <- c ( 'Zhao' , 'Qian' , 'Sun' ) paste( a, 'Ming' ) [ 1 ] "Zhao Ming" "Qian Ming" "Sun Ming" substr( x = month.name , start = 1 , stop = 3 ) [ 1 ] "Jan" "Feb" "Mar" "Apr" "May" "Jun" "Jul" "Aug" "Sep" "Oct" "Nov" "Dec" toupper( month.name ) [ 1 ] "JANUARY" "FEBRUARY" "MARCH" "APRIL" "MAY" "JUNE" "JULY" [ 8 ] "AUGUST" "SEPTEMBER" "OCTOBER" "NOVEMBER" "DECEMBER" tolower( month.name ) [ 1 ] "january" "february" "march" "april" "may" "june" "july" [ 8 ] "august" "september" "october" "november" "december" gsub( ) a <- c ( 'Zhao' , 'Qian' , 'Sun' , 'Li' ) grep( 'Sun' , a) [ 1 ] 3 grep( 'a' , a) [ 1 ] 1 2 grep( 'ao' , a) [ 1 ] 1 grep( 'au' , a) integer( 0 ) a <- c ( 'A+' , 'AC' ) grep( 'A+' , a, fixed = T ) [ 1 ] 1 grep( 'A+' , a, fixed = F ) [ 1 ] 1 2 match( 'A' , a) [ 1 ] NA match( 'A+' , a) [ 1 ] 1 match( 'AC' , a) [ 1 ] 2 strsplit( x = 'hello my dear.' , split = ' ' ) [[ 1 ] ] [ 1 ] "hello" "my" "dear." a <- c ( 'oh my god!' , 'hello my dear.' ) strsplit( a, ' ' ) [[ 1 ] ] [ 1 ] "oh" "my" "god!" [[ 2 ] ] [ 1 ] "hello" "my" "dear." a [ 1 ] 1 2 3 4 5 6 7 8 9 10 11 12 13 b [ 1 ] "heitao" "fangpian" "hongtao" "meihua" outer( b, a, FUN = paste) [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ , 6 ] [ 1 , ] "heitao 1" "heitao 2" "heitao 3" "heitao 4" "heitao 5" "heitao 6" [ 2 , ] "fangpian 1" "fangpian 2" "fangpian 3" "fangpian 4" "fangpian 5" "fangpian 6" [ 3 , ] "hongtao 1" "hongtao 2" "hongtao 3" "hongtao 4" "hongtao 5" "hongtao 6" [ 4 , ] "meihua 1" "meihua 2" "meihua 3" "meihua 4" "meihua 5" "meihua 6" [ , 7 ] [ , 8 ] [ , 9 ] [ , 10 ] [ , 11 ] [ , 12 ] [ 1 , ] "heitao 7" "heitao 8" "heitao 9" "heitao 10" "heitao 11" "heitao 12" [ 2 , ] "fangpian 7" "fangpian 8" "fangpian 9" "fangpian 10" "fangpian 11" "fangpian 12" [ 3 , ] "hongtao 7" "hongtao 8" "hongtao 9" "hongtao 10" "hongtao 11" "hongtao 12" [ 4 , ] "meihua 7" "meihua 8" "meihua 9" "meihua 10" "meihua 11" "meihua 12" [ , 13 ] [ 1 , ] "heitao 13" [ 2 , ] "fangpian 13" [ 3 , ] "hongtao 13" [ 4 , ] "meihua 13" outer( b, a, FUN = paste, sep = '-' )
正则表达式
regular expression regex RE
特点
正则表达式能够凝练字符串的特征
是通用的字符串表达框架
是简洁表达一组字符串的表达式
可以判断某个字符串的特征归属,是否具有这个特征
作用
表达文本类型的特征
同时查找或替换一组字符串
匹配字符串的全部或部分
语法
由字符和操作符构成
基本和常用的正则表达式操作符如下
操作符
说明
例子
.表示任何单个字符
[]给出单个字符的取值范围
[abc]:a或b或c,[a-z]:a-z单个字母
[^]排除此取值范围
[^abc]:非a或b或c
*前一个字符出现>=0次
abc*:ab、abc、abcc、abccc等
+前一个字符出现>=1次
abc+:abc、abcc、abccc等
?前一个字符出现0或1次
abc?:ab、abc
`
`
左右表达式中的一个
{m}前一个字符出现m次
ab{2}c:abbc
{n,}前一个字符出现>=n次
ab{2,}c:abbc、abbbc、abbbbc等
{m,n}前一个字符出现m-n次(含n)
ab{1,3}c:abc、abbc、abbbc
^匹配字符串的开头
^123:字符串以abc开头
$匹配字符串的结尾
123$:字符串以123结尾
()分组标记,内部只能使用`
`
\d数字,等价于[0-9]
\w单词字符,等价于[A-Za-z0-9_]
经典实例
^[A-Za-z]+$:由字母组成的字符串
^[A-Za-z]+$:由字母和数字组成的字符串
^-?[1-9]\d*$:整数
^[1-9]\d*$:正整数
[\u4e00-\u9fa5]:匹配中文字符
日期和时间
时间序列数据
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 class ( sunspots) [ 1 ] "ts" sunspots Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec 1749 58.0 62.6 70.0 55.7 85.0 83.5 94.8 66.3 75.9 75.5 158.6 85.2 1750 73.3 75.9 89.2 88.3 90.0 100.0 85.4 103.0 91.2 65.7 63.3 75.4 1751 70.0 43.5 45.3 56.4 60.7 50.7 66.3 59.8 23.5 23.2 28.5 44.0 ... presidents Qtr1 Qtr2 Qtr3 Qtr4 1945 NA 87 82 75 1946 63 50 43 32 1947 35 60 54 55 ... airmiles Time Series: Start = 1937 End = 1960 Frequency = 1 [ 1 ] 412 480 683 1052 1385 1418 1634 2178 3362 5948 6109 5981 6753 8003 10566 [ 16 ] 12528 14760 16769 19819 22362 25340 25343 29269 30514 class ( airmiles) [ 1 ] "ts" Sys.Date( ) [ 1 ] "2022-01-31" class ( Sys.Date( ) ) [ 1 ] "Date" a <- as.Date( '2022-02-02' , format = '%Y-%m-%d' ) class ( a) [ 1 ] "Date" ? strftime seq( as.Date( '2022-01-01' ) , as.Date( '2022-03-01' ) , by= 5 ) [ 1 ] "2022-01-01" "2022-01-06" "2022-01-11" "2022-01-16" "2022-01-21" "2022-01-26" "2022-01-31" [ 8 ] "2022-02-05" "2022-02-10" "2022-02-15" "2022-02-20" "2022-02-25" runif( 10 , min = 0 , max = 10 ) [ 1 ] 9.0326531 0.5515827 6.8552402 0.8496375 7.1430260 5.6753642 9.7609197 7.2243469 5.2854405 [ 10 ] 7.4681394 sales <- round ( runif( 48 , min = 50 , max = 100 ) , 0 ) sales [ 1 ] 98 96 77 51 55 54 80 71 59 65 70 81 54 75 64 99 74 58 89 93 78 80 83 71 53 99 69 64 91 94 [ 31 ] 72 54 85 83 58 74 55 76 54 57 87 79 83 72 54 87 96 61 ts( sales, start = c ( 2022 , 1 ) , end = c ( 2022 , 12 ) , frequency = 12 ) Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec 2022 98 96 77 51 55 54 80 71 59 65 70 81 ts( sales, start = c ( 2022 , 1 ) , end = c ( 2025 , 1 ) , frequency = 1 ) Time Series: Start = 2022 End = 2025 Frequency = 1 [ 1 ] 98 96 77 51 ts( sales, start = c ( 2022 , 1 ) , end = c ( 2023 , 1 ) , frequency = 12 ) Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov Dec 2022 98 96 77 51 55 54 80 71 59 65 70 81 2023 54 ts( sales, start = c ( 2022 , 1 ) , end = c ( 2023 , 1 ) , frequency = 4 ) Qtr1 Qtr2 Qtr3 Qtr4 2022 98 96 77 51 2023 55
数据处理
数据获取
获取数据的途径:
键盘直接敲入
读取存储在外部文件中已预处理好的数据
访问数据库系统
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 patientID <- c ( 1 , 2 , 3 , 4 ) admdate <- c ( "10/15/2009" , "11/01/2009" , "10/21/2009" , "10/28/2009" ) age <- c ( 25 , 34 , 28 , 52 ) diabetes <- c ( "Type1" , "Type2" , "Type1" , "Type1" ) status <- c ( "Poor" , "Improved" , "Excellent" , "Poor" ) data <- data.frame( patientID, age, diabetes, status) data2 <- data.frame( patientID= character( ) , admdate= character( ) , age= numeric( ) , diabetes= character( ) , status= character( ) ) data2 <- edit( data2) fix( data2) install.packages( "RODBC" )
文件读取
纯文本文件.csv .txt
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 read.table( '/home/lah/Desktop/R_DATA/input.txt' ) Ozone Solar.R Wind Temp Month Day 1 41 190 7.4 67 5 1 2 36 118 8.0 72 5 2 3 12 149 12.6 74 5 3 ... x <- read.table( '/home/lah/Desktop/R_DATA/input.csv' ) x V1 V2 1 , "mpg" , "cyl" , "disp" , "hp" , "drat" , "wt" , "qsec" , "vs" , "am" , "gear" , "carb" 2 Mazda RX4 , 21 , 6 , 160 , 110 , 3.9 , 2.62 , 16.46 , 0 , 1 , 4 , 4 3 Mazda RX4 Wag , 21 , 6 , 160 , 110 , 3.9 , 2.875 , 17.02 , 0 , 1 , 4 , 4 ... x <- read.table( '/home/lah/Desktop/R_DATA/input.csv' , sep = ',' ) head( x) V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 1 mpg cyl disp hp drat wt qsec vs am gear carb2 Mazda RX4 21 6 160 110 3.9 2.62 16.46 0 1 4 4 3 Mazda RX4 Wag 21 6 160 110 3.9 2.875 17.02 0 1 4 4 ... x <- read.table( '/home/lah/Desktop/R_DATA/input.csv' , sep = ',' , header = T , row.names = 1 ) head( x) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1 x <- read.table( '/home/lah/Desktop/R_DATA/input.csv' , sep = ',' , skip = 30 ) x V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 1 Ferrari Dino 19.7 6 145 175 3.62 2.77 15.5 0 1 5 6 2 Maserati Bora 15.0 8 301 335 3.54 3.57 14.6 0 1 5 8 3 Volvo 142 E 21.4 4 121 109 4.11 2.78 18.6 1 1 4 2 x <- read.table( '/home/lah/Desktop/R_DATA/input.csv' , sep = ',' , nrows = 2 ) x V1 V2 V3 V4 V5 V6 V7 V8 V9 V10 V11 V12 1 mpg cyl disp hp drat wt qsec vs am gear carb2 Mazda RX4 21 6 160 110 3.9 2.62 16.46 0 1 4 4 read.fwf( '/home/lah/Desktop/R_DATA/fwf.txt' , widths = c ( 11 , 11 ) ) read.csv( file, header = TRUE , sep = "," , quote = "\"" , dec = "." , fill = TRUE , comment.char = "" , ...) read.csv2( file, header = TRUE , sep = ";" , quote = "\"" , dec = "," , fill = TRUE , comment.char = "" , ...) read.delim( file, header = TRUE , sep = "\t" , quote = "\"" , dec = "." , fill = TRUE , comment.char = "" , ...) read.delim2( file, header = TRUE , sep = "\t" , quote = "\"" , dec = "," , fill = TRUE , comment.char = "" , ...)
网页表格,剪贴板,压缩文件
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 x <- read.table( "https://codeload.github.com/mperdeck/LINQtoCSV/zip/master" , header = TRUE ) library( XML) readHTMLTable( "https://en.wikipedia.org/wiki/World_population" , which= 3 ) RSiteSearch( 'manhattan' ) A search query has been submitted to http: / / search.r- project.org 计算结果应很快就在瀏覽器里打开 read.table( 'clipboard' , header = T ) read.table( gzfile( '/home/lah/Desktop/R_DATA/newfile.txt.gz' ) ) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 ...
scan()和readLines()
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 readLines( '/home/lah/Desktop/R_DATA/newfile.txt' , n= 4 ) [ 1 ] "\"Ozone\" \"Solar.R\" \"Wind\" \"Temp\" \"Month\" \"Day\"" [ 2 ] "\"1\" 41 190 7.4 67 5 1" [ 3 ] "\"2\" 36 118 8 72 5 2" [ 4 ] "\"3\" 12 149 12.6 74 5 3" readLines( '/home/lah/Desktop/R_DATA/scan.txt' ) [ 1 ] "one\t2\t3\tfour\t5\t6" "one\t2\t3\tfour\t5\t6" "one\t2\t3\tfour\t5\t6" [ 4 ] "one\t2\t3\tfour\t5\t6" "one\t2\t3\tfour\t5\t6" "one\t2\t3\tfour\t5\t6" [ 7 ] "one\t2\t3\tfour\t5\t6" "one\t2\t3\tfour\t5\t6" "one\t2\t3\tfour\t5\t6" [ 10 ] "one\t2\t3\tfour\t5\t6" "" read.table( '/home/lah/Desktop/R_DATA/scan.txt' , sep = '\t' ) V1 V2 V3 V4 V5 V6 1 one 2 3 four 5 6 2 one 2 3 four 5 6 3 one 2 3 four 5 6 4 one 2 3 four 5 6 5 one 2 3 four 5 6 6 one 2 3 four 5 6 7 one 2 3 four 5 6 8 one 2 3 four 5 6 9 one 2 3 four 5 6 10 one 2 3 four 5 6 scan( '/home/lah/Desktop/R_DATA/scan.txt' , what = list ( character( 3 ) ) ) Read 60 records [[ 1 ] ] [ 1 ] "one" "2" "3" "four" "5" "6" "one" "2" "3" "four" "5" "6" "one" [ 14 ] "2" "3" "four" "5" "6" "one" "2" "3" "four" "5" "6" "one" "2" [ 27 ] "3" "four" "5" "6" "one" "2" "3" "four" "5" "6" "one" "2" "3" [ 40 ] "four" "5" "6" "one" "2" "3" "four" "5" "6" "one" "2" "3" "four" [ 53 ] "5" "6" "one" "2" "3" "four" "5" "6" scan( '/home/lah/Desktop/R_DATA/scan.txt' , what = list ( character( 3 ) , numeric( 1 ) , numeric( 1 ) ) ) Read 20 records [[ 1 ] ] [ 1 ] "one" "four" "one" "four" "one" "four" "one" "four" "one" "four" "one" "four" "one" [ 14 ] "four" "one" "four" "one" "four" "one" "four" [[ 2 ] ] [ 1 ] 2 5 2 5 2 5 2 5 2 5 2 5 2 5 2 5 2 5 2 5 [[ 3 ] ] [ 1 ] 3 6 3 6 3 6 3 6 3 6 3 6 3 6 3 6 3 6 3 6 scan( '/home/lah/Desktop/R_DATA/scan.txt' , what = list ( A= character( 3 ) , B= numeric( 1 ) , C= numeric( 1 ) , D= character( 3 ) , E= numeric( 1 ) , F = numeric( 1 ) ) ) Read 10 records $ A [ 1 ] "one" "one" "one" "one" "one" "one" "one" "one" "one" "one" $ B [ 1 ] 2 2 2 2 2 2 2 2 2 2 $ C [ 1 ] 3 3 3 3 3 3 3 3 3 3 $ D [ 1 ] "four" "four" "four" "four" "four" "four" "four" "four" "four" "four" $ E [ 1 ] 5 5 5 5 5 5 5 5 5 5 $ F [ 1 ] 6 6 6 6 6 6 6 6 6 6
文件写入
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 write( x, file = '2.txt' ) write( x, file = '2.txt' , ncolumns = 10 ) write( x, file = '2.txt' , ncolumns = 10 , sep = '\t' ) write.table( state.x77, '2.txt' ) write.table( mtcars, '2.txt' ) write.table( mtcars, '2.csv' , sep = ',' ) write.table( mtcars, '2.csv' , append = T ) write.table( mtcars, '2.csv' , quote = F ) write.table( mtcars, '2.csv' , row.names = F ) write.table( mtcars, '2.csv' , col.names = F ) write.table( mtcars, gzfile( '1.txt.gz' ) ) library( foreign) help( package = 'foreign' )
Excel文件读写
XLConnect包(需Java:https://blog.csdn.net/weixin_36350581/article/details/114214756)运行失败
XLSX包 运行失败
openxlsx包
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 read.xlsx( 'writeXLSX2.xlsx' ) read.xlsx( 'writeXLSX2.xlsx' , sheet = 'state info' ) read.xlsx( 'writeXLSX2.xlsx' , startRow = 48 ) read.xlsx( 'writeXLSX2.xlsx' , colNames = F ) read.xlsx( 'writeXLSX2.xlsx' , rowNames = F ) read.xlsx( 'writeXLSX2.xlsx' , skipEmptyRows = F ) read.xlsx( 'writeXLSX2.xlsx' , skipEmptyCols = F ) read.xlsx( 'writeXLSX2.xlsx' , sheet = 'test' , cols = c ( 1 ) ) A 1 1 2 2 3 3 4 4 5 5 6 6 read.xlsx( 'writeXLSX2.xlsx' , sheet = 'test' , cols = c ( 1 : 4 ) ) A B C 1 1 2 3 2 2 3 4 3 3 4 5 4 4 5 6 5 5 6 7 6 6 7 8 read.xlsx( 'writeXLSX2.xlsx' , sheet = 'test' , cols = c ( 1 : 4 ) , rows = c ( 1 : 4 ) ) A B C 1 1 2 3 2 2 3 4 3 3 4 5 write.xlsx( mtcars, 'test.xlsx' ) write.xlsx( mtcars, 'test.xlsx' , asTable = T ) write.xlsx( mtcars, 'test.xlsx' , overwrite = F ) Error in saveWorkbook( wb, file = file, overwrite = overwrite) : File already exists! write.xlsx( mtcars, file = 'car.xlsx' , rowNames= T , colNames= T ) l <- list ( 'Cars Info' = mtcars, 'Iris Info' = iris, 'State info' = data.frame( state.name, state.abb, state.x77) ) write.xlsx( l, 'test.xlsx' )
R 格式文件读写
.rds文件可以保存单个R对象
.rdata文件可以保存对个R对象
存储为R文件有很多优势,特别是对于大数据文件,R会自动压缩处理并包含更多信息。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 head( iris, n = 3 ) Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosasaveRDS( iris, file= 'iris.rds' ) iris_test <- readRDS( "~/R_learn/R_Data_Structure/iris.rds" ) iris_test Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa... save( mtcars, iris, iris3, file = 'test_data.Rdata' ) load( "~/R_learn/R_Data_Structure/test_data.Rdata" ) save.image( file = 'test.Rdata' )
数据转换
数据类型转换
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 head( car, n = 3 ) mpg cyl disp hp drat wt qsec vs am gear carb 1 Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 2 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 3 Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 is.data.frame( car) [ 1 ] TRUE is.matrix ( car) [ 1 ] FALSE is.na ( car) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE Mazda RX4 Wag FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE Datsun 710 FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE ... methods( is) [ 1 ] is.array is.atomic is.call [ 4 ] is.character is.complex is.data.frame [ 7 ] is.double is.element is.empty.model ... dstate.x77 <- as.data.frame( state.x77) View( dstate.x77) is.data.frame( dstate.x77) [ 1 ] TRUE methods( as) [ 1 ] as.array as.array.default as.call [ 4 ] as.character as.character.condition as.character.Date [ 7 ] as.character.default as.character.error as.character.factor ... a <- state.abb head( a) [ 1 ] "AL" "AK" "AZ" "AR" "CA" "CO" dim ( a) <- c ( 5 , 10 ) head( a, n = 3 ) [ , 1 ] [ , 2 ] [ , 3 ] [ , 4 ] [ , 5 ] [ , 6 ] [ , 7 ] [ , 8 ] [ , 9 ] [ , 10 ] [ 1 , ] "AL" "CO" "HI" "KS" "MA" "MT" "NM" "OK" "SD" "VA" [ 2 , ] "AK" "CT" "ID" "KY" "MI" "NE" "NY" "OR" "TN" "WA" [ 3 , ] "AZ" "DE" "IL" "LA" "MN" "NV" "NC" "PA" "TX" "WV" a <- state.abb as.factor( a) [ 1 ] AL AK AZ AR CA CO CT DE FL GA HI ID IL IN IA KS KY LA ME MD MA MI MN MS MO MT NE NV NH NJ [ 31 ] NM NY NC ND OH OK OR PA RI SC SD TN TX UT VT VA WA WV WI WY50 Levels: AK AL AR AZ CA CO CT DE FL GA HI IA ID IL IN KS KY LA MA MD ME MI MN MO MS MT ... WYas.list( a) [[ 1 ] ] [ 1 ] "AL" [[ 2 ] ] [ 1 ] "AK" [[ 3 ] ] [ 1 ] "AZ" ... state <- data.frame( a, state.x77) head( state, n = 3 ) a Population Income Illiteracy Life.Exp Murder HS.Grad Frost Area Alabama AL 3615 3624 2.1 69.05 15.1 41.3 20 50708 Alaska AK 365 6315 1.5 69.31 11.3 66.7 152 566432 Arizona AZ 2212 4530 1.8 70.55 7.8 58.1 15 113417 state$ Population [ 1 ] 3615 365 2212 2110 21198 2541 3100 579 8277 4931 868 813 11197 5313 2861 [ 16 ] 2280 3387 3806 1058 4122 5814 9111 3921 2341 4767 746 1544 590 812 7333 [ 31 ] 1144 18076 5441 637 10735 2715 2284 11860 931 2816 681 4173 12237 1203 472 [ 46 ] 4981 3559 1799 4589 376 b <- state[ 'California' , ] is.data.frame( b) [ 1 ] TRUE unname( b) California CA 21198 5114 1.1 71.71 10.3 62.6 20 156361 head( b) a Population Income Illiteracy Life.Exp Murder HS.Grad Frost Area California CA 21198 5114 1.1 71.71 10.3 62.6 20 156361 unlist( b) a Population Income Illiteracy Life.Exp Murder HS.Grad Frost "CA" "21198" "5114" "1.1" "71.71" "10.3" "62.6" "20" Area "156361" as.vector( b) a Population Income Illiteracy Life.Exp Murder HS.Grad Frost Area California CA 21198 5114 1.1 71.71 10.3 62.6 20 156361 c <- as.vector( b) class ( c ) [ 1 ] "data.frame"
数据取子集、合并、添加
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 state.x77[ 1 : 5 , 1 : 5 ] Population Income Illiteracy Life Exp Murder Alabama 3615 3624 2.1 69.05 15.1 Alaska 365 6315 1.5 69.31 11.3 Arizona 2212 4530 1.8 70.55 7.8 Arkansas 2110 3378 1.9 70.66 10.1 California 21198 5114 1.1 71.71 10.3 state.x77[ c ( 1 , 2 , 3 ) , 1 : 2 ] Population Income Alabama 3615 3624 Alaska 365 6315 Arizona 2212 4530 state.x77[ c ( 1 , 2 , 7 ) , 1 : 2 ] Population Income Alabama 3615 3624 Alaska 365 6315 Connecticut 3100 5348 state.x77[ 1 : 3 , - 1 : - 5 ] HS Grad Frost Area Alabama 41.3 20 50708 Alaska 66.7 152 566432 Arizona 58.1 15 113417 mtcars[ mtcars$ cyl== 4 , ] mpg cyl disp hp drat wt qsec vs am gear carb Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Merc 240 D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2 Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2 ... mtcars[ mtcars$ mpg> 30 , ] mpg cyl disp hp drat wt qsec vs am gear carb Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1 Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2 Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1 Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2 subset( mtcars, mtcars$ mpg> 32 ) mpg cyl disp hp drat wt qsec vs am gear carb Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1 Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1 mtcars[ mtcars$ mpg> 30 & mtcars$ wt> 2 , ] mpg cyl disp hp drat wt qsec vs am gear carb Fiat 128 32.4 4 78.7 66 4.08 2.2 19.47 1 1 4 1 subset( mtcars, mtcars$ mpg> 32 & mtcars$ wt> 2 ) mpg cyl disp hp drat wt qsec vs am gear carb Fiat 128 32.4 4 78.7 66 4.08 2.2 19.47 1 1 4 1 x <- 1: 100 sample( x, 10 ) [ 1 ] 46 29 39 20 12 70 66 74 33 94 sample( x, 10 , replace = T ) [ 1 ] 23 23 22 52 37 20 78 96 34 92 sample( mtcars$ mpg, 5 ) [ 1 ] 22.8 15.2 10.4 21.0 21.0 mtcars[ sample( mtcars$ mpg, 5 ) , ] mpg cyl disp hp drat wt qsec vs am gear carb Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2 Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4 Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1 Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4 Fiat X1- 9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1 mtcars$ mpg <- NULL head( mtcars) cyl disp hp drat wt qsec vs am gear carb Mazda RX4 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 6 160 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 4 108 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 8 360 175 3.15 3.440 17.02 0 0 3 2 Valiant 6 225 105 2.76 3.460 20.22 1 0 3 1 mtcars[ - 1 : - 30 , ] cyl disp hp drat wt qsec vs am gear carb Maserati Bora 8 301 335 3.54 3.57 14.6 0 1 5 8 Volvo 142 E 4 121 109 4.11 2.78 18.6 1 1 4 2 mtcars[ , - 1 : - 8 ] gear carb Mazda RX4 4 4 Mazda RX4 Wag 4 4 Datsun 710 4 1 ... state.abb [ 1 ] "AL" "AK" "AZ" "AR" "CA" "CO" "CT" "DE" "FL" "GA" "HI" "ID" "IL" "IN" "IA" "KS" "KY" "LA" "ME" [ 20 ] "MD" "MA" "MI" "MN" "MS" "MO" "MT" "NE" "NV" "NH" "NJ" "NM" "NY" "NC" "ND" "OH" "OK" "OR" "PA" [ 39 ] "RI" "SC" "SD" "TN" "TX" "UT" "VT" "VA" "WA" "WV" "WI" "WY" data.frame( USArrests, state.abb) Murder Assault UrbanPop Rape state.abb Alabama 13.2 236 58 21.2 AL Alaska 10.0 263 48 44.5 AK Arizona 8.1 294 80 31.0 AZ ... cbind( USArrests, state.abb) Murder Assault UrbanPop Rape state.abb Alabama 13.2 236 58 21.2 AL Alaska 10.0 263 48 44.5 AK Arizona 8.1 294 80 31.0 AZ ... data1 <- head( mtcars, 5 ) data2 <- tail( mtcars, 5 ) rbind( data1, data2) cyl disp hp drat wt qsec vs am gear carb Mazda RX4 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Lotus Europa 4 95.1 113 3.77 1.513 16.90 1 1 5 2 Ford Pantera L 8 351.0 264 4.22 3.170 14.50 0 1 5 4 Ferrari Dino 6 145.0 175 3.62 2.770 15.50 0 1 5 6 Maserati Bora 8 301.0 335 3.54 3.570 14.60 0 1 5 8 Volvo 142 E 4 121.0 109 4.11 2.780 18.60 1 1 4 2 data2 <- head( mtcars, 10 ) rbind( data1, data2) cyl disp hp drat wt qsec vs am gear carb Mazda RX4 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Mazda RX41 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag1 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 7101 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive1 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout1 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Valiant 6 225.0 105 2.76 3.460 20.22 1 0 3 1 Duster 360 8 360.0 245 3.21 3.570 15.84 0 0 3 4 Merc 240 D 4 146.7 62 3.69 3.190 20.00 1 0 4 2 Merc 230 4 140.8 95 3.92 3.150 22.90 1 0 4 2 Merc 280 6 167.6 123 3.92 3.440 18.30 1 0 4 4 data3 <- rbind( data1, data2) duplicated( data3) [ 1 ] FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE data3[ duplicated( data3) , ] cyl disp hp drat wt qsec vs am gear carb Mazda RX41 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag1 6 160 110 3.90 2.875 17.02 0 1 4 4 Datsun 7101 4 108 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive1 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout1 8 360 175 3.15 3.440 17.02 0 0 3 2 data3[ ! duplicated( data3) , ] cyl disp hp drat wt qsec vs am gear carb Mazda RX4 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Valiant 6 225.0 105 2.76 3.460 20.22 1 0 3 1 Duster 360 8 360.0 245 3.21 3.570 15.84 0 0 3 4 Merc 240 D 4 146.7 62 3.69 3.190 20.00 1 0 4 2 Merc 230 4 140.8 95 3.92 3.150 22.90 1 0 4 2 Merc 280 6 167.6 123 3.92 3.440 18.30 1 0 4 4 unique( data3) cyl disp hp drat wt qsec vs am gear carb Mazda RX4 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Valiant 6 225.0 105 2.76 3.460 20.22 1 0 3 1 Duster 360 8 360.0 245 3.21 3.570 15.84 0 0 3 4 Merc 240 D 4 146.7 62 3.69 3.190 20.00 1 0 4 2 Merc 230 4 140.8 95 3.92 3.150 22.90 1 0 4 2 Merc 280 6 167.6 123 3.92 3.440 18.30 1 0 4 4
数据翻转、更改、排序
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 t( mtcars[ 1 : 3 , 1 : 3 ] ) Mazda RX4 Mazda RX4 Wag Datsun 710 mpg 21 21 22.8 cyl 6 6 4.0 disp 160 160 108.0 letters [ 1 ] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m" "n" "o" "p" "q" "r" "s" "t" "u" "v" "w" "x" [ 25 ] "y" "z" rev( letters ) [ 1 ] "z" "y" "x" "w" "v" "u" "t" "s" "r" "q" "p" "o" "n" "m" "l" "k" "j" "i" "h" "g" "f" "e" "d" "c" [ 25 ] "b" "a" women height weight 1 58 115 2 59 117 3 60 120 ... rev( row.names( women) ) [ 1 ] "15" "14" "13" "12" "11" "10" "9" "8" "7" "6" "5" "4" "3" "2" "1" women[ rev( row.names( women) ) , ] height weight 15 72 164 14 71 159 13 70 154 ... women$ height* 2.54 [ 1 ] 147.32 149.86 152.40 154.94 157.48 160.02 162.56 165.10 167.64 170.18 172.72 175.26 177.80 [ 14 ] 180.34 182.88 data.frame( weight= women$ height* 2.54 , women$ weight) weight women.weight 1 147.32 115 2 149.86 117 3 152.40 120 ... transform( women, height= height* 2.54 ) height weight 1 147.32 115 2 149.86 117 3 152.40 120 ... transform( women, cm= height* 2.54 ) height weight cm 1 58 115 147.32 2 59 117 149.86 3 60 120 152.40 ... rivers [ 1 ] 735 320 325 392 524 450 1459 135 465 600 330 336 280 315 870 906 202 329 290 [ 20 ] 1000 600 505 1450 840 1243 890 350 407 286 280 525 720 390 250 327 230 265 850 [ 39 ] 210 630 260 230 360 730 600 306 390 420 291 710 340 217 281 352 259 250 470 [ 58 ] 680 570 350 300 560 900 625 332 2348 1171 3710 2315 2533 780 280 410 460 260 255 [ 77 ] 431 350 760 618 338 981 1306 500 696 605 250 411 1054 735 233 435 490 310 460 [ 96 ] 383 375 1270 545 445 1885 380 300 380 377 425 276 210 800 420 350 360 538 1100 [ 115 ] 1205 314 237 610 360 540 1038 424 310 300 444 301 268 620 215 652 900 525 246 [ 134 ] 360 529 500 720 270 430 671 1770 rev( rivers) [ 1 ] 1770 671 430 270 720 500 529 360 246 525 900 652 215 620 268 301 444 300 310 [ 20 ] 424 1038 540 360 610 237 314 1205 1100 538 360 350 420 800 210 276 425 377 380 [ 39 ] 300 380 1885 445 545 1270 375 383 460 310 490 435 233 735 1054 411 250 605 696 [ 58 ] 500 1306 981 338 618 760 350 431 255 260 460 410 280 780 2533 2315 3710 1171 2348 [ 77 ] 332 625 900 560 300 350 570 680 470 250 259 352 281 217 340 710 291 420 390 [ 96 ] 306 600 730 360 230 260 630 210 850 265 230 327 250 390 720 525 280 286 407 [ 115 ] 350 890 1243 840 1450 505 600 1000 290 329 202 906 870 315 280 336 330 600 465 [ 134 ] 135 1459 450 524 392 325 320 735 sort( rivers) [ 1 ] 135 202 210 210 215 217 230 230 233 237 246 250 250 250 255 259 260 260 265 [ 20 ] 268 270 276 280 280 280 281 286 290 291 300 300 300 301 306 310 310 314 315 [ 39 ] 320 325 327 329 330 332 336 338 340 350 350 350 350 352 360 360 360 360 375 [ 58 ] 377 380 380 383 390 390 392 407 410 411 420 420 424 425 430 431 435 444 445 [ 77 ] 450 460 460 465 470 490 500 500 505 524 525 525 529 538 540 545 560 570 600 [ 96 ] 600 600 605 610 618 620 625 630 652 671 680 696 710 720 720 730 735 735 760 [ 115 ] 780 800 840 850 870 890 900 900 906 981 1000 1038 1054 1100 1171 1205 1243 1270 1306 [ 134 ] 1450 1459 1770 1885 2315 2348 2533 3710 sort( rivers, decreasing = T ) [ 1 ] 3710 2533 2348 2315 1885 1770 1459 1450 1306 1270 1243 1205 1171 1100 1054 1038 1000 981 906 [ 20 ] 900 900 890 870 850 840 800 780 760 735 735 730 720 720 710 696 680 671 652 [ 39 ] 630 625 620 618 610 605 600 600 600 570 560 545 540 538 529 525 525 524 505 [ 58 ] 500 500 490 470 465 460 460 450 445 444 435 431 430 425 424 420 420 411 410 [ 77 ] 407 392 390 390 383 380 380 377 375 360 360 360 360 352 350 350 350 350 340 [ 96 ] 338 336 332 330 329 327 325 320 315 314 310 310 306 301 300 300 300 291 290 [ 115 ] 286 281 280 280 280 276 270 268 265 260 260 259 255 250 250 250 246 237 233 [ 134 ] 230 230 217 215 210 210 202 135 sort( mtcars) Error in `[.data.frame`( x, order( x, na.last = na.last, decreasing = decreasing) ) : 选择了未定义的列 In addition: Warning message: In xtfrm.data.frame( x) : cannot xtfrm data frames mtcars[ sort( row.names( mtcars) ) , ] mpg cyl disp hp drat wt qsec vs am gear carb AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2 Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4 Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4 Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4 ... rivers [ 1 ] 735 320 325 392 524 450 1459 135 465 600 330 336 280 315 870 906 202 329 290 [ 20 ] 1000 600 505 1450 840 1243 890 350 407 286 280 525 720 390 250 327 230 265 850 [ 39 ] 210 630 260 230 360 730 600 306 390 420 291 710 340 217 281 352 259 250 470 ... sort( rivers) [ 1 ] 135 202 210 210 215 217 230 230 233 237 246 250 250 250 255 259 260 260 265 [ 20 ] 268 270 276 280 280 280 281 286 290 291 300 300 300 301 306 310 310 314 315 [ 39 ] 320 325 327 329 330 332 336 338 340 350 350 350 350 352 360 360 360 360 375 ... order( rivers) [ 1 ] 8 17 39 108 129 52 36 42 91 117 133 34 56 87 76 55 41 75 37 127 138 107 13 30 [ 25 ] 72 53 29 19 49 61 103 124 126 46 94 123 116 14 2 3 35 18 11 65 12 81 51 27 [ 49 ] 60 78 111 54 43 112 119 134 97 105 102 104 96 33 47 4 28 73 88 48 110 122 106 139 ... rivers[ 8 ] [ 1 ] 135 rivers[ 17 ] [ 1 ] 202 order( - rivers) [ 1 ] 68 70 66 69 101 141 7 23 83 98 25 115 67 114 89 121 20 82 16 63 131 26 15 38 [ 25 ] 24 109 71 79 1 90 44 32 137 50 85 58 140 130 40 64 128 80 118 86 10 21 45 59 [ 49 ] 62 99 120 113 135 31 132 5 22 84 136 93 57 9 74 95 6 100 125 92 77 139 106 122 rivers[ 68 ] [ 1 ] 3710 sort( mtcars$ mpg) [ 1 ] 10.4 10.4 13.3 14.3 14.7 15.0 15.2 15.2 15.5 15.8 16.4 17.3 17.8 18.1 18.7 19.2 19.2 19.7 21.0 [ 20 ] 21.0 21.4 21.4 21.5 22.8 22.8 24.4 26.0 27.3 30.4 30.4 32.4 33.9 order( mtcars$ mpg) [ 1 ] 15 16 24 7 17 31 14 23 22 29 12 13 11 6 5 10 25 30 1 2 4 32 21 3 9 8 27 26 19 28 18 20 mtcars[ order( mtcars$ mpg) , ] mpg cyl disp hp drat wt qsec vs am gear carb Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4 Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4 Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4 Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4 ... mtcars[ order( mtcars$ mpg, mtcars$ disp) , ] mpg cyl disp hp drat wt qsec vs am gear carb Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4 Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4 Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4 Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4 ... order( - rivers) [ 1 ] 68 70 66 69 101 141 7 23 83 98 25 115 67 114 89 121 20 82 16 63 131 26 15 38 [ 25 ] 24 109 71 79 1 90 44 32 137 50 85 58 140 130 40 64 128 80 118 86 10 21 45 59 [ 49 ] 62 99 120 113 135 31 132 5 22 84 136 93 57 9 74 95 6 100 125 92 77 139 106 122
数据框内数据计算
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 wp <- as.data.frame( WorldPhones) head( wp) N.Amer Europe Asia S.Amer Oceania Africa Mid.Amer 1951 45939 21574 2876 1815 1646 89 555 1956 60423 29990 4708 2568 2366 1411 733 1957 64721 32510 5230 2695 2526 1546 773 ... rowSums( wp) 1951 1956 1957 1958 1959 1960 1961 74494 102199 110001 118399 124801 133709 141700 colMeans( wp) N.Amer Europe Asia S.Amer Oceania Africa Mid.Amer 66747.5714 34343.4286 6229.2857 2772.2857 2625.0000 1484.0000 841.7143 cbind( wp, Total= rowSums( wp) ) N.Amer Europe Asia S.Amer Oceania Africa Mid.Amer Total 1951 45939 21574 2876 1815 1646 89 555 74494 1956 60423 29990 4708 2568 2366 1411 733 102199 1957 64721 32510 5230 2695 2526 1546 773 110001 ... new <- cbind( wp, Total= rowSums( wp) ) rbind( new, Mean= colMeans( new) ) N.Amer Europe Asia S.Amer Oceania Africa Mid.Amer Total 1951 45939.00 21574.00 2876.000 1815.000 1646 89 555.0000 74494.0 1956 60423.00 29990.00 4708.000 2568.000 2366 1411 733.0000 102199.0 1957 64721.00 32510.00 5230.000 2695.000 2526 1546 773.0000 110001.0 1958 68484.00 35218.00 6662.000 2845.000 2691 1663 836.0000 118399.0 1959 71799.00 37598.00 6856.000 3000.000 2868 1769 911.0000 124801.0 1960 76036.00 40341.00 8220.000 3145.000 3054 1905 1008.0000 133709.0 1961 79831.00 43173.00 9053.000 3338.000 3224 2005 1076.0000 141700.0 Mean 66747.57 34343.43 6229.286 2772.286 2625 1484 841.7143 115043.3 apply( wp, 1 , FUN = sum ) 1951 1956 1957 1958 1959 1960 1961 74494 102199 110001 118399 124801 133709 141700 apply( wp, 2 , sum ) N.Amer Europe Asia S.Amer Oceania Africa Mid.Amer 467233 240404 43605 19406 18375 10388 5892 class ( apply( wp, 2 , sum ) ) [ 1 ] "numeric" apply( wp, 1 , sd) 1951 1956 1957 1958 1959 1960 1961 17309.22 22712.46 24362.16 25790.60 27116.58 28712.04 30213.56 lapply( state.center, length ) $ x[ 1 ] 50 $ y[ 1 ] 50 sapply( state.center, length ) x y 50 50 tapply( state.name, state.division, length ) New England Middle Atlantic South Atlantic East South Central West South Central 6 3 8 4 4 East North Central West North Central Mountain Pacific 5 7 8 5 x <- c ( 1 , 2 , 3 , 4 , 5 ) x- mean( x) [ 1 ] - 2 - 1 0 1 2 sd( x) [ 1 ] 1.581139 ( x- mean( x) ) / sd( x) [ 1 ] - 1.2649111 - 0.6324555 0.0000000 0.6324555 1.2649111 scale( x, center = T , scale = F ) [ , 1 ] [ 1 , ] - 2 [ 2 , ] - 1 [ 3 , ] 0 [ 4 , ] 1 [ 5 , ] 2 attr ( , "scaled:center" ) [ 1 ] 3 scale( x, center = T , scale = T ) [ , 1 ] [ 1 , ] - 1.2649111 [ 2 , ] - 0.6324555 [ 3 , ] 0.0000000 [ 4 , ] 0.6324555 [ 5 , ] 1.2649111 attr ( , "scaled:center" ) [ 1 ] 3 attr ( , "scaled:scale" ) [ 1 ] 1.581139 heatmap( scale( state.x77, T , T ) )
reshape2包应用tidyr包应用作用:整理数据,合并数据,长宽数据互转
gather():宽数据→长数据(建议使用更新的pivot_longer()和pivot_wider())
spread():长数据→宽数据
unite():多列合并为一列
seperate():一列拆分为多列
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 td <- mtcars[ 1 : 2 , 1 : 3 ] td mpg cyl disp Mazda RX4 21.0 6 160.0 Mazda RX4 Wag 21.0 6 160.0 td <- data.frame( car= rownames( td) , td) td car mpg cyl disp Mazda RX4 Mazda RX4 21.0 6 160.0 Mazda RX4 Wag Mazda RX4 Wag 21.0 6 160.0 gather( td, key = 'Type' , value = 'Value' , mpg, cyl, disp) car Type Value 1 Mazda RX4 mpg 21 2 Mazda RX4 Wag mpg 21 3 Mazda RX4 cyl 6 4 Mazda RX4 Wag cyl 6 5 Mazda RX4 disp 160 6 Mazda RX4 Wag disp 160 gather( td, key = 'Type' , value = 'Value' , mpg: disp) car Type Value 1 Mazda RX4 mpg 21 2 Mazda RX4 Wag mpg 21 3 Mazda RX4 cyl 6 4 Mazda RX4 Wag cyl 6 5 Mazda RX4 disp 160 6 Mazda RX4 Wag disp 160 gather( td, key = 'Type' , value = 'Value' , mpg: disp, - cyl) car cyl Type Value 1 Mazda RX4 6 mpg 21 2 Mazda RX4 Wag 6 mpg 21 3 Mazda RX4 6 disp 160 4 Mazda RX4 Wag 6 disp 160 gather( td, key = 'Type' , value = 'Value' , 3 : 4 ) car mpg Type Value 1 Mazda RX4 21 cyl 6 2 Mazda RX4 Wag 21 cyl 6 3 Mazda RX4 21 disp 160 4 Mazda RX4 Wag 21 disp 160 gtd <- gather( td, key = 'Type' , value = 'Value' , mpg: disp, - cyl) gtd car cyl Type Value 1 Mazda RX4 6 mpg 21 2 Mazda RX4 Wag 6 mpg 21 3 Mazda RX4 6 disp 160 4 Mazda RX4 Wag 6 disp 160 spread( gtd, key = 'Type' , value = 'Value' ) car cyl disp mpg 1 Mazda RX4 6 160 21 2 Mazda RX4 Wag 6 160 21 x X 1 < NA > 2 1 - 2 3 2 - 3 4 3 - 4 separate( x, col = X, into = c ( 'A' , 'B' ) ) A B 1 < NA < NA > 2 1 2 3 2 3 4 3 4 separate( x, col = X, into = c ( 'A' , 'B' ) , sep = '-' ) A B 1 < NA < NA > 2 1 2 3 2 3 4 3 4 x A B 1 < NA < NA > 2 1 2 3 2 3 4 3 4 unite( x, col= X) X 1 NA_NA2 1 _23 2 _34 3 _4unite( x, col= X, sep = '-' ) X 1 NA - NA 2 1 - 2 3 2 - 3 4 3 - 4
dplyr包应用1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 > dplyr:: filter( mtcars, mpg> 30 ) mpg cyl disp hp drat wt qsec vs am gear carb model Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1 Fiat 128 Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2 Honda Civic Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1 Toyota Corolla Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2 Lotus Europa > dplyr:: filter( mtcars, mpg> 30 & wt> 2 ) mpg cyl disp hp drat wt qsec vs am gear carb model Fiat 128 32.4 4 78.7 66 4.08 2.2 19.47 1 1 4 1 Fiat 128 > dplyr:: distinct( rbind( mtcars[ 1 : 2 , ] , mtcars[ 1 : 3 , ] ) ) mpg cyl disp hp drat wt qsec vs am gear carb model Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 Mazda RX4 Wag Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 Datsun 710 > dplyr:: slice( mtcars, 1 : 3 ) mpg cyl disp hp drat wt qsec vs am gear carb model Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 Mazda RX4 Wag Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 Datsun 710 > dplyr:: sample_n( mtcars, 3 ) mpg cyl disp hp drat wt qsec vs am gear carb model Porsche 914 - 2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2 Porsche 914 - 2 Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1 Fiat 128 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Mazda RX4 Wag > length ( row.names( mtcars) ) [ 1 ] 32 > dplyr:: sample_frac( mtcars, 0.1 ) mpg cyl disp hp drat wt qsec vs am gear carb model Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.5 0 1 5 6 Ferrari Dino AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.3 0 0 3 2 AMC Javelin Merc 450 SE 16.4 8 275.8 180 3.07 4.070 17.4 0 0 3 3 Merc 450 SE > dplyr:: arrange( mtcars, mpg) mpg cyl disp hp drat wt qsec vs am gear carb model Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4 Cadillac Fleetwood Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4 Lincoln Continental Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4 Camaro Z28 ... > dplyr:: arrange( mtcars, - mpg) > dplyr:: arrange( mtcars, desc( mpg) ) mpg cyl disp hp drat wt qsec vs am gear carb model Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1 Toyota Corolla Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1 Fiat 128 Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2 Honda Civic ... > summarise( mtcars, avg= mean( mpg) ) avg 1 20.09062 > summarise( mtcars, avg= mean( cyl) ) avg 1 6.1875 > summarise( mtcars, sum = sum ( mpg) ) sum 1 642.9 > mtcars %>% summarise( sum = sum ( mpg) ) sum 1 642.9 > head( mtcars, 10 ) %>% tail( 3 ) mpg cyl disp hp drat wt qsec vs am gear carb Merc 240 D 24.4 4 146.7 62 3.69 3.19 20.0 1 0 4 2 Merc 230 22.8 4 140.8 95 3.92 3.15 22.9 1 0 4 2 Merc 280 19.2 6 167.6 123 3.92 3.44 18.3 1 0 4 4 > dplyr:: group_by( mtcars, cyl) mpg cyl disp hp drat wt qsec vs am gear carb < dbl> < dbl> < dbl> < dbl> < dbl> < dbl> < dbl> < dbl> < dbl> < dbl> < dbl> 1 21 6 160 110 3.9 2.62 16.5 0 1 4 4 2 21 6 160 110 3.9 2.88 17.0 0 1 4 4 3 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1 4 21.4 6 258 110 3.08 3.22 19.4 1 0 3 1 5 18.7 8 360 175 3.15 3.44 17.0 0 0 3 2 6 18.1 6 225 105 2.76 3.46 20.2 1 0 3 1 7 14.3 8 360 245 3.21 3.57 15.8 0 0 3 4 8 24.4 4 147. 62 3.69 3.19 20 1 0 4 2 9 22.8 4 141. 95 3.92 3.15 22.9 1 0 4 2 10 19.2 6 168. 123 3.92 3.44 18.3 1 0 4 4 > dplyr:: group_by( mtcars, cyl) %>% summarise( ) cyl < dbl> 1 4 2 6 3 8 > dplyr:: group_by( mtcars, cyl) %>% summarise( ave= mean( mpg) ) cyl ave < dbl> < dbl> 1 4 26.7 2 6 19.7 3 8 15.1 > dplyr:: group_by( mtcars, cyl) %>% summarise( ave= mean( mpg) ) %>% arrange( ave) cyl ave < dbl> < dbl> 1 8 15.1 2 6 19.7 3 4 26.7 > mt <- mutate( mtcars, new = mpg+ disp) > head( mt, 3 ) mpg cyl disp hp drat wt qsec vs am gear carb new Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 181.0 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 181.0 Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 130.8 > a x1 x2 1 A 1 2 B 2 3 C 3 > b x1 x3 1 A TRUE 2 B FALSE 3 D TRUE > dplyr:: left_join( a, b, by= "x1" ) x1 x2 x3 1 A 1 TRUE 2 B 2 FALSE 3 C 3 NA > dplyr:: right_join( a, b, 'x1' ) x1 x2 x3 1 A 1 TRUE 2 B 2 FALSE 3 D NA TRUE > inner_join( a, b, 'x1' ) x1 x2 x3 1 A 1 TRUE 2 B 2 FALSE > dplyr:: full_join( a, b, by= "x1" ) x1 x2 x3 1 A 1 TRUE 2 B 2 FALSE 3 C 3 NA 4 D NA TRUE > dplyr:: semi_join( a, b, by= "x1" ) x1 x2 1 A 1 2 B 2 > dplyr:: anti_join( a, b, by= "x1" ) x1 x2 1 C 3 > car1 mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 > car2 mpg cyl disp hp drat wt qsec vs am gear carb Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1 Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4 Merc 240 D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2 > intersect( car1, car2) mpg cyl disp hp drat wt qsec vs am gear carb Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 > union( car1, car2) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1 Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4 Merc 240 D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2 > union_all( car1, car2) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 Datsun 710. ..3 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive...4 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout...5 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Datsun 710. ..6 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive...7 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout...8 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1 Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4 Merc 240 D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2 > setdiff( car1, car2) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21 6 160 110 3.9 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21 6 160 110 3.9 2.875 17.02 0 1 4 4 > setdiff( car2, car1) mpg cyl disp hp drat wt qsec vs am gear carb Valiant 18.1 6 225.0 105 2.76 3.46 20.22 1 0 3 1 Duster 360 14.3 8 360.0 245 3.21 3.57 15.84 0 0 3 4 Merc 240 D 24.4 4 146.7 62 3.69 3.19 20.00 1 0 4 2
R函数
函数的选项参数
函数选项类型
输入控制
输出控制
调节
数学统计函数
d概率密度函数
p分布函数
q分布函数的反函数
r随机数函数
如正态分布函数norm()加上这4个前缀:
dnorm(), pnorm(), qnorm(), rnorm()
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 > rnorm( n= 10 , mean= 5 , sd= 1 ) [ 1 ] 6.329543 4.232249 6.033233 4.837938 3.984797 5.299808 5.202550 2.070615 5.830317 5.936402 > round ( rnorm( n= 10 , mean= 5 , sd= 1 ) ) [ 1 ] 5 4 5 5 5 4 4 5 6 3 > runif( 5 ) [ 1 ] 0.04366431 0.34873894 0.29974298 0.39224632 0.08980883 > runif( 5 , 0 , 10 ) [ 1 ] 0.83081773 2.22285093 0.07499167 4.59838033 6.37444314 > runif( 5 , min = 0 , max = 10 ) [ 1 ] 4.5102458 0.3494494 9.4862655 4.6722139 0.5983668 > round ( runif( 5 , min = 0 , max = 10 ) ) [ 1 ] 1 5 8 6 10 > set.seed( 10 ) > round ( runif( 5 , min = 0 , max = 10 ) ) [ 1 ] 5 3 4 7 1 > round ( runif( 5 , min = 0 , max = 10 ) ) [ 1 ] 2 3 3 6 4 > round ( runif( 5 , min = 0 , max = 10 ) ) [ 1 ] 7 6 1 6 4 > set.seed( 10 ) > round ( runif( 5 , min = 0 , max = 10 ) ) [ 1 ] 5 3 4 7 1
描述性统计函数
频数统计函数
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 > table( mtcars$ cyl) 4 6 8 11 7 14 > prop.table( table( mtcars$ cyl) ) 4 6 8 0.34375 0.21875 0.43750 > split( mtcars, mtcars$ cyl) $ `4 ` mpg cyl disp hp drat wt qsec vs am gear carb Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2 $`6` mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4 $`8` mpg cyl disp hp drat wt qsec vs am gear carb Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2 Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4 > split(mtcars, mtcars$cyl)['4'] $`4` mpg cyl disp hp drat wt qsec vs am gear carb Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1 Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2 # 计算连续值频数和频率 > range(mtcars$mpg) [1] 10.4 33.9 > seq(10, 40, 5) [1] 10 15 20 25 30 35 40 > cut(mtcars$mpg, seq(10, 35, 5)) [1] (20,25] (20,25] (20,25] (20,25] (15,20] (15,20] (10,15] (20,25] (20,25] (15,20] (15,20] (15,20] [13] (15,20] (15,20] (10,15] (10,15] (10,15] (30,35] (30,35] (30,35] (20,25] (15,20] (15,20] (10,15] [25] (15,20] (25,30] (25,30] (30,35] (15,20] (15,20] (10,15] (20,25] Levels: (10,15] (15,20] (20,25] (25,30] (30,35] > table(cut(mtcars$mpg, seq(10, 35, 5))) (10,15] (15,20] (20,25] (25,30] (30,35] 6 12 8 2 4 > prop.table(table(cut(mtcars$mpg, seq(10, 35, 5)))) (10,15] (15,20] (20,25] (25,30] (30,35] 0.1875 0.3750 0.2500 0.0625 0.1250 # 二维,talbe() xtabs() > library(vcd) > Arthritis ID Treatment Sex Age Improved 1 57 Treated Male 27 Some 2 46 Treated Male 29 None 3 77 Treated Male 30 None > table(Arthritis$Treatment, Arthritis$Improved) None Some Marked Placebo 29 7 7 Treated 13 7 21 > xtabs(~ Treatment + Improved, data = Arthritis) Improved Treatment None Some Marked Placebo 29 7 7 Treated 13 7 21 > with(data = Arthritis, {table(Treatment, Improved)}) Improved Treatment None Some Marked Placebo 29 7 7 Treated 13 7 21 # 边际频数及统计,1表示按行统计,2表示按列统计 # margin.table() addmargins() > x <- xtabs(~ Treatment + Improved, data = Arthritis) > margin.table(x) [1] 84 > margin.table(x, 1) Treatment Placebo Treated 43 41 > margin.table(x, 2) Improved None Some Marked 42 14 28 > prop.table(x, 1) Improved Treatment None Some Marked Placebo 0.6744186 0.1627907 0.1627907 Treated 0.3170732 0.1707317 0.5121951 > prop.table(x, 2) Improved Treatment None Some Marked Placebo 0.6904762 0.5000000 0.2500000 Treated 0.3095238 0.5000000 0.7500000 > prop.table(x) Improved Treatment None Some Marked Placebo 0.34523810 0.08333333 0.08333333 Treated 0.15476190 0.08333333 0.25000000 > addmargins(x) Improved Treatment None Some Marked Sum Placebo 29 7 7 43 Treated 13 7 21 41 Sum 42 14 28 84 > addmargins(x, 1) Improved Treatment None Some Marked Placebo 29 7 7 Treated 13 7 21 Sum 42 14 28 > addmargins(x, 2) Improved Treatment None Some Marked Sum Placebo 29 7 7 43 Treated 13 7 21 41 # 三维ftable() > y <- xtabs(~ Treatment + Improved + Sex, data = Arthritis) > y , , Sex = Female Improved Treatment None Some Marked Placebo 19 7 6 Treated 6 5 16 , , Sex = Male Improved Treatment None Some Marked Placebo 10 0 1 Treated 7 2 5 > ftable(y) Sex Female Male Treatment Improved Placebo None 19 10 Some 7 0 Marked 6 1 Treated None 6 7 Some 5 2 Marked 16 5
独立性检验函数
独立性检验是根据频数信息判断两类因子彼此相关或者独立的假设检验。
独立性检验算法:
卡方检验
Fisher检验
Cochran-Mantel-Haenszel检验
假设检验(Hypothesis Testing):由抽样判断总体。
原假设:没有发生;
备择假设:发生了
p value:原假设为真时,得到最大的或者超出所得到的的检验统计计算量的概率
p值一般定到0.05,当p<0.05时,拒绝原假设,p>0.05时,不拒绝原假设(根据情况调整p值)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 > Arthritis ID Treatment Sex Age Improved 1 57 Treated Male 27 Some2 46 Treated Male 29 None3 77 Treated Male 30 None> table( Arthritis$ Treatment, Arthritis$ Improved) None Some Marked Placebo 29 7 7 Treated 13 7 21 > mytable <- table( Arthritis$ Treatment, Arthritis$ Improved) > chisq.test( mytable) Pearson's Chi-squared test data: mytable X-squared = 13.055, df = 2, p-value = 0.001463 # p>0.05,不拒绝原假设,Sex和Improved独立 > mytable <- table(Arthritis$Sex, Arthritis$Improved) > chisq.test(mytable) Pearson' s Chi- squared testdata: mytable X- squared = 4.8407 , df = 2 , p- value = 0.08889 Warning message: In chisq.test( mytable) : Chi- squared approximation may be incorrect > mytable <- table( Arthritis$ Treatment, Arthritis$ Improved) > fisher.test( mytable) Fisher's Exact Test for Count Data data: mytable p-value = 0.001393 alternative hypothesis: two.sided # Cochran-Mantel-Haenszel检验,p<0.05,拒绝原假设,Treatment和Improved有关 > mytable <- xtabs(~ Treatment+Improved+Sex, data = Arthritis) > mytable , , Sex = Female Improved Treatment None Some Marked Placebo 19 7 6 Treated 6 5 16 , , Sex = Male Improved Treatment None Some Marked Placebo 10 0 1 Treated 7 2 5 > mantelhaen.test(mytable) Cochran-Mantel-Haenszel test data: mytable Cochran-Mantel-Haenszel M^2 = 14.632, df = 2, p-value = 0.0006647 # 顺序很重要 > mytable <- xtabs(~ Treatment+Sex+Improved, data = Arthritis) > mantelhaen.test(mytable) Mantel-Haenszel chi-squared test with continuity correction data: mytable Mantel-Haenszel X-squared = 2.0863, df = 1, p-value = 0.1486 alternative hypothesis: true common odds ratio is not equal to 1 95 percent confidence interval: 0.8566711 8.0070521 sample estimates: common odds ratio 2.619048 > mytable <- xtabs(~ Sex+Improved+Treatment, data = Arthritis) > mantelhaen.test(mytable) Cochran-Mantel-Haenszel test data: mytable Cochran-Mantel-Haenszel M^2 = 6.846, df = 2, p-value = 0.03261
相关性分析函数
相关系数:
Pearson相关系数
Spearman相关系数
Kendall相关系数
偏相关系数
多分格相关系数(polychoric)
多系列相关系数(polyserial)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 > x <- state.x77[ , c ( 1 , 2 , 3 , 6 ) ] > cor( x) Population Income Illiteracy HS Grad Population 1.00000000 0.2082276 0.1076224 - 0.09848975 Income 0.20822756 1.0000000 - 0.4370752 0.61993232 Illiteracy 0.10762237 - 0.4370752 1.0000000 - 0.65718861 HS Grad - 0.09848975 0.6199323 - 0.6571886 1.00000000 > y <- state.x77[ , c ( 4 , 5 ) ] > cor( y) Life Exp Murder Life Exp 1.0000000 - 0.7808458 Murder - 0.7808458 1.0000000 > cor( x, y) Life Exp Murder Population - 0.06805195 0.3436428 Income 0.34025534 - 0.2300776 Illiteracy - 0.58847793 0.7029752 HS Grad 0.58221620 - 0.4879710 > cov( x) Population Income Illiteracy HS Grad Population 19931683.759 571229.780 292.8679592 - 3551.509551 Income 571229.780 377573.306 - 163.7020408 3076.768980 Illiteracy 292.868 - 163.702 0.3715306 - 3.235469 HS Grad - 3551.510 3076.769 - 3.2354694 65.237894 > library( ggm) > colnames( state.x77) > pcor( c ( 1 , 5 , 2 , 3 , 6 ) , cov( state.x77) ) [ 1 ] 0.3462724
相关性检验函数
相关性检验函数cor.test(),给出p值和相关系数
1 2 3 4 5 6 7 8 9 10 11 12 > cor.test( state.x77[ , 'Murder' ] , state.x77[ , 'Illiteracy' ] ) Pearson's product-moment correlation data: state.x77[, "Murder"] and state.x77[, "Illiteracy"] t = 6.8479, df = 48, p-value = 1.258e-08 alternative hypothesis: true correlation is not equal to 0 95 percent confidence interval: 0.5279280 0.8207295 sample estimates: cor 0.7029752
psych包的corr.test()函数可计算多个变量的相关性,并给出p值
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 > library( psych) > corr.test( x, y) Call: corr.test( x = x, y = y) Correlation matrix Life Exp Murder Population - 0.07 0.34 Income 0.34 - 0.23 Illiteracy - 0.59 0.70 HS Grad 0.58 - 0.49 Sample Size [ 1 ] 50 These are the unadjusted probability values. The probability values adjusted for multiple tests are in the p.adj object. Life Exp Murder Population 0.64 0.01 Income 0.02 0.11 Illiteracy 0.00 0.00 HS Grad 0.00 0.00 To see confidence intervals of the correlations, print with the short= FALSE option
偏相关性检验
1 2 3 4 5 6 7 8 9 10 11 12 13 > library( ggm) > x <- pcor( c ( 1 , 5 , 2 , 3 , 6 ) , cov( state.x77) ) > x[ 1 ] 0.3462724 > pcor.test( x, 3 , 50 ) $ tval[ 1 ] 2.476049 $ df[ 1 ] 45 $ pvalue[ 1 ] 0.01711252
Student’s t test
1 2 3 4 5 6 7 8 9 10 11 12 13 14 > library( MASS) > UScrime> t.test( Prob ~ So, data = UScrime) Welch Two Sample t- test data: Prob by So t = - 3.8954 , df = 24.925 , p- value = 0.0006506 alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0 95 percent confidence interval: - 0.03852569 - 0.01187439 sample estimates: mean in group 0 mean in group 1 0.03851265 0.06371269
绘图函数
见第三章
自定义函数
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 myfun <- function ( x, na.omit= FALSE ) { if ( na.omit) x <- x[ ! is.na ( x) ] m <- mean( x) n <- length ( x) s <- sd( x) skew <- sum ( x- m^ 3 / s^ 3 ) / n kurt <- sum ( x- m^ 4 / s^ 4 ) / n- 3 return ( c ( n= m, mean= m, stdev= s, skew= skew, kurtosis= kurt) ) } > for ( i in 1 : 10 ) { print( '11' ) } > i= 1 > while ( i<= 10 ) { print( '2' ) ;i= i+ 1 } > ifelse( i> 10 , print( 'yes' ) , print( 'No' ) )
数据分析实战
线性回归
回归(Regression):用用预测变量(自变量、解释变量)来预测响应变量(因变量、结果变量)
回归
用途
简单线性
用一个量化的解释变量预测一个量化的响应变量
多项式
用一个量化的解释变量预测一个量化的响应变量,模型的关系是n阶多项式
多元线性
用两个或多个量化的解释变量预测一个量化的响应变量
多变量
用一个或多个解释变量预测多个响应变量
Logistic
用一个或多个解释变量预测一个类别型响应变量
泊松
用一个或多个解释变量预测一个代表频数的响应变量
Cox比例风险
用一个或多个解释变量预测一个事件 ( 死亡、失败或旧病复发)发生的时间
时间序列
对误差项相关的时间序列数据建模
非线性
用一个或多个量化的解释变量预测一个量化的响应变量,不过模型是非线性的
非参数
用一个或多个量化的解释变量预测一个量化的响应变量,模型的形式源自数据形式,不事先设定
稳健
用一个或多个量化的解释变量预测一个量化的响应变量,能抵御强影响点的干扰
线性回归:
简单线性回归
多项式
多元线性
多变量
Logistic
简单线性回归
普通最小二乘回归法(OLS)
lm()函数进行线性回归
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 > women height weight 1 58 115 2 59 117 3 60 120 > fit <- lm( weight ~ height, data = women) > fitCall: lm( formula = weight ~ height, data = women) Coefficients: ( Intercept) height - 87.52 3.45 > summary( fit) Call: lm( formula = weight ~ height, data = women) Residuals: Min 1 Q Median 3 Q Max - 1.7333 - 1.1333 - 0.3833 0.7417 3.1167 Coefficients: Estimate Std. Error t value Pr( > | t| ) ( Intercept) - 87.51667 5.93694 - 14.74 1.71e-09 * * * height 3.45000 0.09114 37.85 1.09e-14 * * * - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 1.525 on 13 degrees of freedom Multiple R- squared: 0.991 , Adjusted R- squared: 0.9903 F - statistic: 1433 on 1 and 13 DF, p- value: 1.091e-14
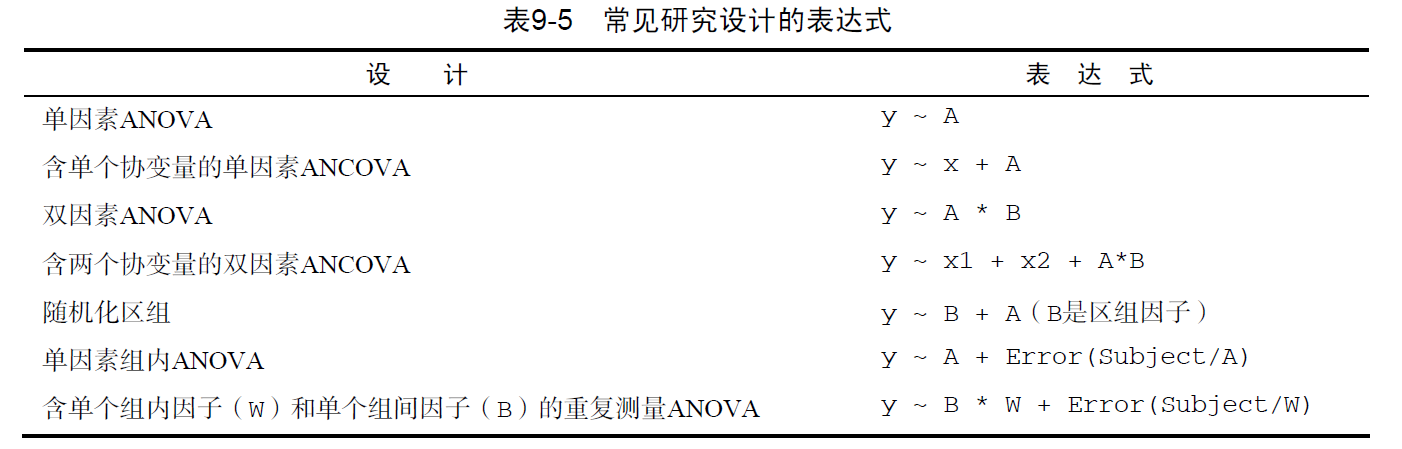
响应变量~预测变量1+预测变量2+预测变量3
拟合线性模型的常用函数
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 > fitCall: lm( formula = weight ~ height, data = women) Coefficients: ( Intercept) height - 87.52 3.45 > coefficients( fit) ( Intercept) height - 87.51667 3.45000 > confint( fit) 2.5 % 97.5 % ( Intercept) - 100.342655 - 74.690679 height 3.253112 3.646888 > confint( fit, level = 0.5 ) 25 % 75 % ( Intercept) - 91.635892 - 83.397441 height 3.386767 3.513233 > fitted( fit) 1 2 3 4 5 6 7 8 9 10 112.5833 116.0333 119.4833 122.9333 126.3833 129.8333 133.2833 136.7333 140.1833 143.6333 11 12 13 14 15 147.0833 150.5333 153.9833 157.4333 160.8833 > residuals( fit) 1 2 3 4 5 6 7 8 2.41666667 0.96666667 0.51666667 0.06666667 - 0.38333333 - 0.83333333 - 1.28333333 - 1.73333333 9 10 11 12 13 14 15 - 1.18333333 - 1.63333333 - 1.08333333 - 0.53333333 0.01666667 1.56666667 3.11666667 > women$ weight- fitted( fit) 1 2 3 4 5 6 7 8 2.41666667 0.96666667 0.51666667 0.06666667 - 0.38333333 - 0.83333333 - 1.28333333 - 1.73333333 9 10 11 12 13 14 15 - 1.18333333 - 1.63333333 - 1.08333333 - 0.53333333 0.01666667 1.56666667 3.11666667 > women1 <- women> predict( fit, women1) 1 2 3 4 5 6 7 8 9 10 112.5833 116.0333 119.4833 122.9333 126.3833 129.8333 133.2833 136.7333 140.1833 143.6333 11 12 13 14 15 147.0833 150.5333 153.9833 157.4333 160.8833 > plot( fit)
多项式回归
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 > fit2 <- lm( weight~ height+ I( height^ 2 ) , data= women) > fit2Call: lm( formula = weight ~ height + I( height^ 2 ) , data = women) Coefficients: ( Intercept) height I( height^ 2 ) 261.87818 - 7.34832 0.08306 > summary( fit2) Call: lm( formula = weight ~ height + I( height^ 2 ) , data = women) Residuals: Min 1 Q Median 3 Q Max - 0.50941 - 0.29611 - 0.00941 0.28615 0.59706 Coefficients: Estimate Std. Error t value Pr( > | t| ) ( Intercept) 261.87818 25.19677 10.393 2.36e-07 * * * height - 7.34832 0.77769 - 9.449 6.58e-07 * * * I( height^ 2 ) 0.08306 0.00598 13.891 9.32e-09 * * * - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 0.3841 on 12 degrees of freedom Multiple R- squared: 0.9995 , Adjusted R- squared: 0.9994 F - statistic: 1.139e+04 on 2 and 12 DF, p- value: < 2.2e-16 > fit3 <- lm( weight~ height+ I( height^ 2 ) + I( height^ 3 ) , data= women) > summary( fit3) Call: lm( formula = weight ~ height + I( height^ 2 ) + I( height^ 3 ) , data = women) Residuals: Min 1 Q Median 3 Q Max - 0.40677 - 0.17391 0.03091 0.12051 0.42191 Coefficients: Estimate Std. Error t value Pr( > | t| ) ( Intercept) - 8.967e+02 2.946e+02 - 3.044 0.01116 * height 4.641e+01 1.366e+01 3.399 0.00594 * * I( height^ 2 ) - 7.462e-01 2.105e-01 - 3.544 0.00460 * * I( height^ 3 ) 4.253e-03 1.079e-03 3.940 0.00231 * * - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 0.2583 on 11 degrees of freedom Multiple R- squared: 0.9998 , Adjusted R- squared: 0.9997 F - statistic: 1.679e+04 on 3 and 11 DF, p- value: < 2.2e-16 > plot( women) > abline( fit) > lines( women$ height, fitted( fit2) , col= 'red' ) > lines( women$ height, fitted( fit3) , col= 'blue' )
多元线性回归
回归系数的含义:其他预测变量保持不变时,一个预测变量增加一个单位后,因变量的增量。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 > state <- as.data.frame( state.x77[ , c ( 'Murder' , 'Life Exp' , 'Illiteracy' , 'Income' , 'HS Grad' ) ] ) > colnames( state) <- c ( 'Murder' , 'Life_Exp' , 'Illiteracy' , 'Income' , 'HS_Grad' ) > fit <- lm( Murder ~ Life_Exp+ Illiteracy+ Income+ HS_Grad, data= state) > fitCall: lm( formula = Murder ~ Life_Exp + Illiteracy + Income + HS_Grad, data = state) Coefficients: ( Intercept) Life_Exp Illiteracy Income HS_Grad 116.507133 - 1.662190 2.786755 0.000718 0.042163 > summary( fit) Call: lm( formula = Murder ~ Life_Exp + Illiteracy + Income + HS_Grad, data = state) Residuals: Min 1 Q Median 3 Q Max - 3.715 - 1.524 0.113 1.737 3.848 Coefficients: Estimate Std. Error t value Pr( > | t| ) ( Intercept) 1.165e+02 1.976e+01 5.897 4.44e-07 * * * Life_Exp - 1.662e+00 2.822e-01 - 5.891 4.53e-07 * * * Illiteracy 2.787e+00 6.708e-01 4.154 0.000144 * * * Income 7.180e-04 6.024e-04 1.192 0.239535 HS_Grad 4.216e-02 5.736e-02 0.735 0.466095 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 2.028 on 45 degrees of freedom Multiple R- squared: 0.7229 , Adjusted R- squared: 0.6983 F - statistic: 29.35 on 4 and 45 DF, p- value: 4.961e-12 > coef( fit) ( Intercept) Life_Exp Illiteracy Income HS_Grad 1.165071e+02 - 1.662190e+00 2.786755e+00 7.179837e-04 4.216343e-02 > fit1 <- lm( Murder ~ Life_Exp+ Illiteracy+ Income+ HS_Grad, data= state) > fit2 <- lm( Murder ~ Life_Exp+ Illiteracy, data= state) > summary( fit2) Call: lm( formula = Murder ~ Life_Exp + Illiteracy, data = state) Residuals: Min 1 Q Median 3 Q Max - 4.3725 - 1.5867 0.0284 1.1919 4.3641 Coefficients: Estimate Std. Error t value Pr( > | t| ) ( Intercept) 114.2162 19.6698 5.807 5.27e-07 * * * Life_Exp - 1.5446 0.2716 - 5.688 7.96e-07 * * * Illiteracy 2.2557 0.5981 3.772 0.000453 * * * - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 2.063 on 47 degrees of freedom Multiple R- squared: 0.7004 , Adjusted R- squared: 0.6876 F - statistic: 54.94 on 2 and 47 DF, p- value: 4.998e-13 > AIC( fit1, fit2) df AIC fit1 6 219.3144 fit2 4 219.2232
有交互项的多元线性回归
有时候变量之间不是独立的,有交互关系,如车辆马力与车重有关系.
若两个预测变量的交互项显著,说明响应变量与其中一个预测变量的关系依赖于另一个预测变量的水平。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 > fit <- lm( mpg~ hp+ wt+ hp: wt, data= mtcars) > summary( fit) Call: lm( formula = mpg ~ hp + wt + hp: wt, data = mtcars) Residuals: Min 1 Q Median 3 Q Max - 3.0632 - 1.6491 - 0.7362 1.4211 4.5513 Coefficients: Estimate Std. Error t value Pr( > | t| ) ( Intercept) 49.80842 3.60516 13.816 5.01e-14 * * * hp - 0.12010 0.02470 - 4.863 4.04e-05 * * * wt - 8.21662 1.26971 - 6.471 5.20e-07 * * * hp: wt 0.02785 0.00742 3.753 0.000811 * * * - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 2.153 on 28 degrees of freedom Multiple R- squared: 0.8848 , Adjusted R- squared: 0.8724 F - statistic: 71.66 on 3 and 28 DF, p- value: 2.981e-13
AIC()只能比较两个模型
如果变量很多,可以采用逐步回归法和全子集回归法
逐步回归法:模型会一次添加或删除变量,向前或向后逐步回归
全子集回归法:计算所有模型后选出最优模型,更全面
逐步回归法
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 > library( MASS) > states <- as.data.frame( state.x77[ , c ( "Murder" , "Population" , + "Illiteracy" , "Income" , "Frost" ) ] ) > fit <- lm( Murder ~ Population + Illiteracy + Income + Frost, + data= states) > stepAIC( fit, direction = "backward" ) Start: AIC= 97.75 Murder ~ Population + Illiteracy + Income + Frost Df Sum of Sq RSS AIC - Frost 1 0.021 289.19 95.753 - Income 1 0.057 289.22 95.759 < none> 289.17 97.749 - Population 1 39.238 328.41 102.111 - Illiteracy 1 144.264 433.43 115.986 Step: AIC= 95.75 Murder ~ Population + Illiteracy + Income Df Sum of Sq RSS AIC - Income 1 0.057 289.25 93.763 < none> 289.19 95.753 - Population 1 43.658 332.85 100.783 - Illiteracy 1 236.196 525.38 123.605 Step: AIC= 93.76 Murder ~ Population + Illiteracy Df Sum of Sq RSS AIC < none> 289.25 93.763 - Population 1 48.517 337.76 99.516 - Illiteracy 1 299.646 588.89 127.311 Call: lm( formula = Murder ~ Population + Illiteracy, data = states) Coefficients: ( Intercept) Population Illiteracy 1.6515497 0.0002242 4.0807366
全子集回归法
1 2 3 4 5 6 > library( leaps) Warning message: > states <- as.data.frame( state.x77[ , c ( "Murder" , "Population" , "Illiteracy" , "Income" , "Frost" ) ] ) > leaps <- regsubsets( Murder ~ Population + Illiteracy + Income + Frost, data= states, nbest= 4 ) > plot( leaps, scale= "adjr2" )
回归诊断
模型是否最佳?是否经得起更多数据的检验?
满足OLS模型的统计假设才能够用lm()进行拟合
正态性:对于固定的自变量值,因变量成正态分布
独立性:因变量之间相互独立
线性:因变量和自变量之间为线性关系
同方差性:因变量的方差不随自变量的水平不同而变化。也称不变方差
plot(fit)的四幅图即是对这四个条件的反应
1 2 3 4 5 opar <- par( no.readonly= TRUE ) fit <- lm( weight ~ height, data= women) par( mfrow= c ( 2 , 2 ) ) plot( fit) par( opar)
残差——拟合图
ANOVA
Analysis of Variance,ANOVA,也称变异数分析,一种统计检验,用于分析两个以上 样本之间的均值的差异。
方差分析分类:
单因素方差分析ANOVA(组内、组间)
双因素方差分析ANOVA
协方差分析ANCOVA
多元方差分析MANOVA
多元协方差分析MANCOVA
aov()和lm()均可进行方差分析,aov()用法类似
常见的公式
公式中各项的顺序很重要。
one-way ANOVA
探究单个单个因素(自变量)对响应变量的影响。
one-way ANOVA,单因素方差分析,一个自变量(independent variable)
适用情况: 自变量至少有3个水平(level,即类型为因子factor),即至少3个不同组或类别,因变量(dependent variable)为数值型(numeric)
ANOVA可检测因变量是否随自变量的水平变化而变化,比如
自变量为微信使用频率,分为低、中、高三个组,探究其晚上睡眠时长是否有差异。
自变量为汽水品牌,收集可口可乐、百事、芬达、康师傅的数据,分析其每100 mL汽水的价格是否有差异。
自变量为肥料种类,使用3种肥料对作物施肥,分析作物产量是否有差异。
ANOVA的零假设(null hypothesis,H0 ):不同组之间的均值没有差异
备择假设(alternative hypothesis, Ha ):至少有一个组与因变量的总体均值显著不同
原理:计算各组的均值是否与因变量总体均值不同,如果有任何一个组的均值与总体均值显著不同,则拒绝零假设。ANOVA使用F检验(F-test)进行统计的显著性分析。由于误差是针对整个组比较计算的,所以F检验允许一次比较多个均值,而不是针对每个单独的双向比较(t-test)。F检验将每组均值的方差与总体组的方差进行比较,如果组内方差小于组间方差,则F检验得到的F值很大,因此观察到的差异是真实存在的可能性更高,而不是偶然现象。
ANOVA假设条件
观测之间相互独立:使用有效的统计学方法收集数据,观测之间没有隐藏的联系
响应变量符合正态分布:因变量的值符合正态分布
同方差性:被比较的每组的方差是相似的,如果组间方差不同,则方差分析可能不适合
实例
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 > oneway <- read.csv( 'crop.data.csv' , colClasses = c ( 'factor' , 'factor' , 'factor' , 'numeric' ) ) > head( oneway) density block fertilizer yield 1 1 1 1 177.2287 2 2 2 1 177.5500 3 1 3 1 176.4085 4 2 4 1 177.7036 5 1 1 1 177.1255 6 2 2 1 176.7783 > summary( oneway) density block fertilizer yield 1 : 48 1 : 24 1 : 32 Min. : 175.4 2 : 48 2 : 24 2 : 32 1 st Qu.: 176.5 3 : 24 3 : 32 Median : 177.1 4 : 24 Mean : 177.0 3 rd Qu.: 177.4 Max. : 179.1 > fit <- aov( yield~ fertilizer, data = oneway) > summary( fit) Df Sum Sq Mean Sq F value Pr( > F ) fertilizer 2 6.07 3.0340 7.863 7e-04 * * * Residuals 93 35.89 0.3859 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1
ANOVA 输出内容解释
第一列:自变量和模型残差(model residuals),也即模型误差(model error)
Df:为自变量的自由度(自变量level-1)和残差的自由度(观测总数-1-????),fertilizer自由度为3-1=2,残差为96-1-2=93Sum Sq:由该变量解释的组均值和总体均值之间的平方和(sum of squares),fertilizer平方和为6.07,残差为35.89Mean Sq:平方和的均值,即平方和÷自由度F value:来自F检验(F-test)的检验统计量,每个自变量的平均方差÷残差的平均方差,F值越大,与自变量相关的变化越有可能是真实的,而不是偶然的Pr(>F):F统计量的p值,表示,如果组均值之间没有差异的零假设为真时,F值发生的可能性
由于自变量fertilizer的p < 0.05,显著,拒绝零假设,所以肥料种类对作物产量有显著影响。
ANOVA 能够分析自变量的水平之间是否存在差异,但哪些差异是显著的却不知道。
可以使用TukeyHSD()函数进行 Tukey’s Honestly-Significant Difference post-hoc test
Tukey test 对组进行两两比较,并使用保守误差(conservative error estimate)估计找出在统计上不同的组
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 > TukeyHSD( fit) Tukey multiple comparisons of means 95 % family- wise confidence level Fit: aov( formula = yield ~ fertilizer, data = oneway) $ fertilizer diff lwr upr p adj 2 - 1 0.1761687 - 0.19371896 0.5460564 0.4954705 3 - 1 0.5991256 0.22923789 0.9690133 0.0006125 3 - 2 0.4229569 0.05306916 0.7928445 0.0208735 > aggregate( oneway$ yield, by= list ( oneway$ fertilizer) , FUN= mean) Group.1 x 1 1 176.7570 2 2 176.9332 3 3 177.3562
输出内容解释
Fit:检验的模型
diff:两组的均值差
lwr, upr:95%置信区间的上下限
p adj:调整后的p值
结果表明,3-2和3-1之间的差异显著(p < 0.05),肥料3的作物平均产量显著高于肥料1和2;
而肥料1和2之间的作物平均产量差异不显著。
ANOVA 总结方式
We found a statistically-significant difference in average crop yield according to fertilizer type (f(2)=9.073, p < 0.001). A Tukey post-hoc test revealed significant pairwise differences between fertilizer types 3 and 2, with an average difference of 0.42 bushels/acre (p < 0.05) and between fertilizer types 3 and 1, with an average difference of 0.59 bushels/acre (p < 0.01).
multcomp包中的cholesterol数据集
自变量:5种疗法trt
响应变量:问卷得分response
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 > library( multcomp) > head( cholesterol) trt response 1 1 time 3.8612 2 1 time 10.3868 3 1 time 5.9059 4 1 time 3.0609 5 1 time 7.7204 6 1 time 2.7139 > table( cholesterol$ trt) 1 time 2 times 4 times drugD drugE 10 10 10 10 10 > aggregate( cholesterol$ response, by= list ( cholesterol$ trt) , FUN= mean) Group.1 x 1 1 time 5.78197 2 2 times 9.22497 3 4 times 12.37478 4 drugD 15.36117 5 drugE 20.94752 > aggregate( cholesterol$ response, by= list ( cholesterol$ trt) , FUN= sd) Group.1 x 1 1 time 2.878113 2 2 times 3.483054 3 4 times 2.923119 4 drugD 3.454636 5 drugE 3.345003 > fit <- aov( response~ trt, data = cholesterol) > fitCall: aov( formula = response ~ trt, data = cholesterol) Terms: trt Residuals Sum of Squares 1351.3690 468.7504 Deg. of Freedom 4 45 Residual standard error: 3.227488 Estimated effects may be unbalanced > summary( fit) Df Sum Sq Mean Sq F value Pr( > F ) trt 4 1351.4 337.8 32.43 9.82e-13 * * * Residuals 45 468.8 10.4 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 > plot( fit) > fit_lm <- lm( response~ trt, data = cholesterol) > summary( fit_lm) Call: lm( formula = response ~ trt, data = cholesterol) Residuals: Min 1 Q Median 3 Q Max - 6.5418 - 1.9672 - 0.0016 1.8901 6.6008 Coefficients: Estimate Std. Error t value Pr( > | t| ) ( Intercept) 5.782 1.021 5.665 9.78e-07 * * * trt2times 3.443 1.443 2.385 0.0213 * trt4times 6.593 1.443 4.568 3.82e-05 * * * trtdrugD 9.579 1.443 6.637 3.53e-08 * * * trtdrugE 15.166 1.443 10.507 1.08e-13 * * * - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 3.227 on 45 degrees of freedom Multiple R- squared: 0.7425 , Adjusted R- squared: 0.7196 F - statistic: 32.43 on 4 and 45 DF, p- value: 9.819e-13
单因素协方差分析
litter怀孕小鼠数据集
因变量:给药剂量dose,0, 5, 50, 500
响应变量:每窝幼崽平均出生体重weight
协变量:怀孕时间gesttime、产仔数
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 > library( multcomp) > head( litter) dose weight gesttime number 1 0 28.05 22.5 15 2 0 33.33 22.5 14 3 0 36.37 22.0 14 4 0 35.52 22.0 13 5 0 36.77 21.5 15 6 0 29.60 23.0 5 > table( litter$ dose) 0 5 50 500 20 19 18 17 > table( litter$ gesttime) 21.5 22 22.5 23 20 24 27 3 > aggregate( litter$ weight, by= list ( litter$ dose) , FUN= mean) Group.1 x 1 0 32.30850 2 5 29.30842 3 50 29.86611 4 500 29.64647 > fit <- aov( weight~ gesttime+ dose, data = litter) > summary( fit) Df Sum Sq Mean Sq F value Pr( > F ) gesttime 1 134.3 134.30 8.049 0.00597 * * dose 3 137.1 45.71 2.739 0.04988 * Residuals 69 1151.3 16.69 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1
two-way ANOVA
two-way ANOVA,双因素方差分析,两个自变量
ToothGrowth数据集
自变量:喂食种类supp、剂量dose
响应变量:牙齿长度len
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 > head( ToothGrowth) len supp dose 1 4.2 VC 0.5 2 11.5 VC 0.5 3 7.3 VC 0.5 4 5.8 VC 0.5 5 6.4 VC 0.5 6 10.0 VC 0.5 > class ( ToothGrowth$ supp) [ 1 ] "factor" > class ( ToothGrowth$ dose) [ 1 ] "numeric" > ToothGrowth$ dose <- factor( ToothGrowth$ dose) > class ( ToothGrowth$ dose) [ 1 ] "factor" > xtabs( ~ dose+ supp, data = ToothGrowth) supp dose OJ VC 0.5 10 10 1 10 10 2 10 10 > aggregate( ToothGrowth$ len, by= list ( ToothGrowth$ dose, ToothGrowth$ supp) , FUN= mean) Group.1 Group.2 x 1 0.5 OJ 13.23 2 1 OJ 22.70 3 2 OJ 26.06 4 0.5 VC 7.98 5 1 VC 16.77 6 2 VC 26.14 > fit <- aov( len~ supp* dose, data = ToothGrowth) > summary( fit) Df Sum Sq Mean Sq F value Pr( > F ) supp 1 205.4 205.4 15.572 0.000231 * * * dose 2 2426.4 1213.2 92.000 < 2e-16 * * * supp: dose 2 108.3 54.2 4.107 0.021860 * Residuals 54 712.1 13.2 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 > library( HH) > interaction.plot( ToothGrowth$ dose, ToothGrowth$ supp, ToothGrowth$ len)
多元方差分析
当因变量不止一个时,可以使用多元方差分析
MASS包UScereal数据集
自变量:货架shelf
因变量:谷物所含卡路里calories、脂肪fat、糖sugars
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 > library( MASS) > attach( UScereal) > aggregate( calories, fat, sugars, by= list ( shelf) , FUN= mean) Error in mean.default( X[[ i] ] , ...) : 'trim' 必需是长度必需为一的数值 In addition: Warning message: In if ( na.rm) x <- x[ ! is.na ( x) ] : the condition has length > 1 and only the first element will be used > aggregate( cbind( calories, fat, sugars) , by= list ( shelf) , FUN= mean) Group.1 calories fat sugars 1 1 119.4774 0.6621338 6.295493 2 2 129.8162 1.3413488 12.507670 3 3 180.1466 1.9449071 10.856821 > shelf <- factor( shelf) > fit <- manova( cbind( calories, fat, sugars) ~ shelf) > summary( fit) Df Pillai approx F num Df den Df Pr( > F ) shelf 2 0.4021 5.1167 6 122 0.0001015 * * * Residuals 62 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 > summary.aov( fit) Response calories : Df Sum Sq Mean Sq F value Pr( > F ) shelf 2 50435 25217.6 7.8623 0.0009054 * * * Residuals 62 198860 3207.4 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Response fat : Df Sum Sq Mean Sq F value Pr( > F ) shelf 2 18.44 9.2199 3.6828 0.03081 * Residuals 62 155.22 2.5035 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1 Response sugars : Df Sum Sq Mean Sq F value Pr( > F ) shelf 2 381.33 190.667 6.5752 0.002572 * * Residuals 62 1797.87 28.998 - - - Signif. codes: 0 ‘* * * ’ 0.001 ‘* * ’ 0.01 ‘* ’ 0.05 ‘.’ 0.1 ‘ ’ 1
Student’s t test
学生 t 检验(student’s t test,也称 t-test)是一种统计检验,用于比较两组的均值。在假设检验中常用于确定某个过程、处理是否对目标真的有显著影响,或者两组是否有显著差异。
比如:想知道鸢尾花的平均花瓣长度是否和花的种类有关,收集两种鸢尾花的花瓣长度数据,每组测量25次,可使用 t 检验检测两组之间的差异。
零假设 H0 :两组均值之间的真实差异为零,即没有差异
备择假设Ha :真实差异不为零
适用情况: t 检验只能用于比较两组的均值(也称成对比较),如果想比较两组以上,用 ANOVA 检验或 post-hoc test。
t 检验假设
数据之间相互独立
数据满足(或近似满足)正态分布
方差同质性:被比较的组的方差相似
若数据不满足要求,可以尝试非参数检验(nonparametric test),如 Wilcoxon Signed-Rank test。
t 检验类型
one-sample,two-sample,paired t-test?
paired t-test,配对样本均值检验:组来自同一类别(如某种处理的前后测量)
two-sample,两独立样本均值检验:组来自两个不同类别(如两种物种,来自两个城市的人),也即 independent t-stet
one-sample,单样本均值检验:一组和标准值相比较(如将液体的 ph 与中性 ph7 相比)
one-tailed,two-tailed t-test?
two-tailed, 双侧 t 检验:只关注两组是否有差异
one-tailed,单侧 t 检验:需要知道两组的均值谁大谁小
如想知道鸢尾花的平均花瓣长度是否和花的种类有关,观测来自两种花,故使用 two-sample t-test;只关注是否有差异,故使用 two-tailed t-test。
t 检验方程
t = x 1 ˉ − x 2 ˉ s 2 ( 1 n 1 + 1 n 2 ) t=\frac{\bar{x_{1}}-\bar{x_{2}}}{\sqrt{s^{2}(\frac{1}{n_{1} }+ \frac{1}{n_{2} })} }
t = s 2 ( n 1 1 + n 2 1 ) x 1 ˉ − x 2 ˉ
式中,t 为 t 值,x1,x2是两组的均值,s两组的合并标准误差,n1,n2是每组的观测个数
t 值大,则两组的均值差异大于合并标准误差,表明两组的差异显著
实例
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 > flower.data <- read.csv( 'flower.data.csv' , row.names = 1 ) > class ( flower.data$ Species) [ 1 ] "character" > flower.data$ Species <- factor( flower.data$ Species) > class ( flower.data$ Species) [ 1 ] "factor" > head( flower.data) Sepal.Length Sepal.Width Petal.Length Petal.Width Species 1 5.1 3.5 1.4 0.2 setosa2 4.7 3.2 1.3 0.2 setosa3 5.0 3.6 1.4 0.2 setosa4 4.6 3.4 1.4 0.3 setosa5 4.4 2.9 1.4 0.2 setosa6 5.4 3.7 1.5 0.2 setosa> table( flower.data$ Species) setosa virginica 25 25 > t.test( Petal.Length~ Species, flower.data, paired= F , alternative= 'two.sided' ) Welch Two Sample t- test data: Petal.Length by Species t = - 33.719 , df = 30.196 , p- value < 2.2e-16 alternative hypothesis: true difference in means between group setosa and group virginica is not equal to 0 95 percent confidence interval: - 4.331287 - 3.836713 sample estimates: mean in group setosa mean in group virginica 1.456 5.540
t.test()输出内容解释
data:对比项t值:-33.719,虽然是负的,但是大多数情况下,只关注其绝对值df:自由度,30.196,自由度与样本大小有关,表示有多少数据点可用于比较,自由度越大,统计检验越有效p值:2.2e-16,偶然得到这样的t值的概率alternative hypothesis:备择假设,setosa 和 virginica 组的真实均值差异不为095% confidence interval:95% 置信区间,真实的均值差在 95% 可能性下所处的区间,即两组的真实均值差有 95% 可能性处于[-4.331287, -3.836713]区间内;可以更改区间,但 95% 常用mean:各组的均值
由以上结果,样本数据的均值差 = 1.456 - 5.540 = -4.084,置信区间显示均值的真实差异在[-4.331287, -3.836713]之间。所以,95% 的情况下均值的真实差异不为0。p值远小于 0.05,所以可以拒绝原假设(均值差异为0),即可以说,均值的真实差异显著不为0。
t 检验总结方式
展示重要的信息:t 值,p 值,自由度
也可以带上均值(mean)和标准差差(standard deviation)
1 2 3 4 5 6 7 8 > flower.data %>% dplyr:: group_by( Species) %>% dplyr:: summarize( mean_length= mean( Petal.Length) , sd_length= sd( Petal.Length) ) Species mean_length sd_length < fct> < dbl> < dbl> 1 setosa 1.46 0.206 2 virginica 5.54 0.569
功效分析
power analysis,可以帮助在给定置信度的情况下,判断检测到给定效应值所需的样本量。反过来,也可以在给定置信度水平情况下,计算某样本量内能检测到给定效应值的概率。
理论基础
样本大小n:实验设计中每种条件/组中观测的数目
显著性水平p(也称alpha):Type I error probability。也可把他看做是发现效应不发生的概率
功效1-p:1 minus Type II error probability。可把他看成是真实效应发生的概率
效应值ES:在备择假设或研究假设下效应的量。效应值的表达式依赖于假设检验中使用的统计方法
功效分析中研究设计的四个基本量,知三求一
R中使用pwr包进行功效分析,根据不同的假设检验选择不同的函数
线性回归的功效分析:pwr.f2.test()
假设要研究老板风格对员工满意度的影响,是否超过薪水和消费对员工满意度的影响。老板风格用4个变量来估计,薪水和小费与3个变量有关。已有研究表明,薪水和小费能够解释约30%员工满意度的方差,而从现实出发,老板风格至少能解释35%的方差,假定显著性水平为0.05,在90%的置信度水平下,需要多少受试者,才能得到这样的方差贡献率?
1 2 3 4 5 6 7 8 9 10 11 > pwr.f2.test( u= 3 , f2= 0.0769 , sig.level= 0.05 , power= 0.90 ) Multiple regression power calculation u = 3 v = 184.2426 f2 = 0.0769 sig.level = 0.05 power = 0.9
方差分析:pwr.anova.test()
假设2组样品做单因素方差分析,要达到0.9的功效,效应值为0.25,并选择0.05的显著性水平,求每一组的样本量
1 2 3 4 5 6 7 8 9 10 11 12 13 > pwr.anova.test( k= 2 , f= .25 , sig.level= .05 , power= .9 ) Balanced one- way analysis of variance power calculation k = 2 n = 85.03128 f = 0.25 sig.level = 0.05 power = 0.9 NOTE: n is number in each group
t检验:pwr.t.test()
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 > pwr.t.test( d= .8 , sig.level= .05 , power= .9 , type= "two.sample" , alternative= "two.sided" ) Two- sample t test power calculation n = 33.82555 d = 0.8 sig.level = 0.05 power = 0.9 alternative = two.sided NOTE: n is number in * each* group > pwr.t.test( n= 20 , d= .5 , sig.level= .01 , type= "two.sample" , alternative= "two.sided" ) Two- sample t test power calculation n = 20 d = 0.5 sig.level = 0.01 power = 0.1439551 alternative = two.sided NOTE: n is number in * each* group
相关性
1 2 pwr.r.test( r= .25 , sig.level= .05 , power= .90 , alternative= "greater" )
比例检验
1 2 3 pwr.2p.test( h= ES.h( .65 , .6 ) , sig.level= .05 , power= .9 , alternative= "greater" )
卡方检验
1 2 3 4 prob <- matrix( c ( .42 , .28 , .03 , .07 , .10 , .10 ) , byrow= TRUE , nrow= 3 ) ES.w2( prob) pwr.chisq.test( w= .1853 , df= 3 , sig.level= .05 , power= .9 )
广义线性模型
前面所学的线性回归和方差分析基于正态分布的假设
许多变量并不是正态分布
1 2 3 4 5 6 7 8 install.packages( 'robust' ) data( breslow.dat, package= 'robust' ) summary( breslow.dat) fit <- glm( sumY~ Base+ Age+ Trt, data= breslow.dat, family = poisson( link = 'log' ) ) summary( fit) coef( fit) exp ( coef( fit) )
R语言作图
条形图
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 x <- read.csv( '../Rdata/homo_length.csv' ) xx xx <- x[ 1 : 24 , ] xx$ chr xx$ length barplot( height = xx$ length ) barplot( height = xx$ length , names.arg = xx$ chr) barplot( height = xx$ length , names.arg = xx$ chr, las = 2 ) yanse <- sample( colours( ) , 24 , replace = F ) barplot( height = xx$ length , names.arg = xx$ chr, las = 2 , col = yanse) barplot( height = xx$ length , names.arg = xx$ chr, las = 2 , col = yanse, border = F ) barplot( height = xx$ length , names.arg = xx$ chr, las = 3 , col = yanse, border = F , main = 'Human Chromosome Length Distribution' , xlab = 'Chromosome' , ylab = 'Length' )
分组条形图
1 2 3 4 5 6 7 8 9 10 11 12 x <- read.csv( file = '../Rdata/sv_distrubution.csv' , row.names = 1 ) tail( x) head( x) barplot( as.matrix( x) ) barplot( t( as.matrix( x) ) ) barplot( t( as.matrix( x) ) , col = rainbow( 4 ) , beside = T ) colnames( x) barplot( t( as.matrix( x) ) , col = rainbow( 4 ) , legend.text = colnames( x) ) barplot( t( as.matrix( x) ) , col = rainbow( 4 ) , legend.text = colnames( x) , ylim = c ( 0 , 800 ) ) barplot( t( as.matrix( x) ) , col = rainbow( 4 ) , legend.text = colnames( x) , ylim = c ( 0 , 800 ) , main = 'SV Distribution' , ylab = 'Numbers' , xlab = 'Chromosomes' )
直方图
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 x <- read.table( '../Rdata/H37Rv.gff' , sep = '\t' , header = F , skip = 7 , quote = '' ) View( x) head( x$ V3== 'gene' ) [ 1 ] FALSE TRUE FALSE TRUE FALSE TRUE x[ x$ V3== 'gene' , ] x <- x[ x$ V3== 'gene' , ] x <- abs ( x$ V5- x$ V4+ 1 ) length ( x) [ 1 ] 3978 range ( x) [ 1 ] 61 12456 hist( x) hist( x, breaks = 80 ) hist( x, breaks = c ( 0 , 500 , 1000 , 1500 , 2000 , 2500 , 15000 ) ) hist( x, breaks = 80 , freq = F ) hist( x, breaks = 80 , freq = F , density = T ) hist( x, breaks = 80 , freq = F , density = T , col = 'pink' ) hist( x, breaks = 80 , freq = F , col = 'pink' , border = F , main = 'histogram of Gene Length' , xlab = 'Gene Length (bp)' ) h <- hist( x, breaks = 80 , freq = F , col = 'pink' , border = F , main = 'histogram of Gene Length' , xlab = 'Gene Length (bp)' ) class ( h) [ 1 ] "histogram" h$ xname [ 1 ] "x" h$ breaks [ 1 ] 0 200 400 600 800 1000 1200 1400 1600 1800 2000 2200 2400 2600 2800 3000 [ 17 ] 3200 3400 3600 3800 4000 4200 4400 4600 4800 5000 5200 5400 5600 5800 6000 6200 [ 33 ] 6400 6600 6800 7000 7200 7400 7600 7800 8000 8200 8400 8600 8800 9000 9200 9400 [ 49 ] 9600 9800 10000 10200 10400 10600 10800 11000 11200 11400 11600 11800 12000 12200 12400 12600 h$ counts [ 1 ] 120 548 578 601 571 430 356 268 165 84 55 43 27 20 24 12 12 13 7 5 3 5 4 4 [ 25 ] 5 1 1 1 2 0 0 4 1 1 0 0 0 2 0 0 0 0 0 0 0 0 1 1 [ 49 ] 0 1 0 0 0 0 0 1 0 0 0 0 0 0 1 h$ density [ 1 ] 1.508296e-04 6.887883e-04 7.264957e-04 7.554047e-04 7.176973e-04 5.404726e-04 4.474610e-04 [ 8 ] 3.368527e-04 2.073906e-04 1.055807e-04 6.913022e-05 5.404726e-05 3.393665e-05 2.513826e-05 [ 15 ] 3.016591e-05 1.508296e-05 1.508296e-05 1.633987e-05 8.798391e-06 6.284565e-06 3.770739e-06 [ 22 ] 6.284565e-06 5.027652e-06 5.027652e-06 6.284565e-06 1.256913e-06 1.256913e-06 1.256913e-06 [ 29 ] 2.513826e-06 0.000000e+00 0.000000e+00 5.027652e-06 1.256913e-06 1.256913e-06 0.000000e+00 [ 36 ] 0.000000e+00 0.000000e+00 2.513826e-06 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 [ 43 ] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 1.256913e-06 1.256913e-06 0.000000e+00 [ 50 ] 1.256913e-06 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 1.256913e-06 [ 57 ] 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 0.000000e+00 1.256913e-06
散点图
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 m <- read.table( '../Rdata/prok_representative.txt' , sep = '\t' ) View( m) gene_number <- m$ V4 gnome_size <- m$ V2 plot( gnome_size, gene_number, pch= 16 , cex= 0.8 , xlab = 'Gnome Size' , ylab = 'Genes' ) fit <- lm( gene_number ~ gnome_size) fit Call: lm( formula = gene_number ~ gnome_size) Coefficients: ( Intercept) gnome_size 286.6 843.7 abline( fit, col= 'red' ) text( 12 , 6000 , labels= 'y=843.691x+286.598\nR2=0.97' ) c [ 1 ] 0.9676204 round ( summary( fit) $ adj.r.squared, 2 ) [ 1 ] 0.97 rr <- round ( summary( fit) $ adj.r.squared, 2 ) intercept <- round ( summary( fit) $ coefficients[ 1 ] , 2 ) intercept [ 1 ] 286.6 slope <- round ( summary( fit) $ coefficients[ 2 ] , 2 ) slope [ 1 ] 843.69 eq <- bquote( atop( "y = " * .( slope) * " x + " * .( intercept) , R^ 2 == .( rr) ) ) text( 12 , 6000 , eq)
饼图
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 read.table( '../Rdata/Species.txt' ) V1 V2 V3 1 Viruses noname 31.42034 2 Bacteria Bacteroidetes 31.27472 3 Bacteria Fibrobacteres 24.74666 4 Bacteria Firmicutes 8.06092 5 Bacteria Spirochaetes 2.32585 6 Archaea Euryarchaeota 1.42253 7 Bacteria Proteobacteria 0.74899 x <- read.table( '../Rdata/Species.txt' ) pie( x$ V3) lab <- paste( x$ V2, '\n' , x$ V3, '%' ) lab [ 1 ] "noname \n 31.42034 %" "Bacteroidetes \n 31.27472 %" "Fibrobacteres \n 24.74666 %" [ 4 ] "Firmicutes \n 8.06092 %" "Spirochaetes \n 2.32585 %" "Euryarchaeota \n 1.42253 %" [ 7 ] "Proteobacteria \n 0.74899 %" pie( x$ V3, labels = lab, col = rainbow( nrow( x) ) ) pie( x$ V3, labels = lab, col = rainbow( nrow( x) ) , radius = 1 , cex = 0.8 ) install.packages( "plotrix" ) library( plotrix) s pie3D( x$ V3, labels = lab, col = rainbow( nrow( x) ) , radius = 1 , cex = 0.8 ) pie3D( x$ V3, labels = lab, col = rainbow( nrow( x) ) , radius = 1 , cex = 0.8 , explode = 0.2 ) pieplot3d <- pie( x$ V3, col = rainbow( nrow( x) ) , radius = 1 , explode = 0.1 ) pie3D.labels( pieplot3d, labels = lab, labelcex = 0.8 , height = 0.3 , labelrad = 1.4 ) pieplot3d <- pie( x$ V3, labels = '' , col = rainbow( nrow( x) ) , radius = 1 ) ang <- floating.pie( 0 , 0 , x$ V3) pie.labels( x= 0 , y= 0 , angles = ang, labels= lab, radius= 1.2 , minangle = 0.17 )
箱线图
箱线图展示了数据的5个指标(最小值、下四分位数、中位数、上四分位数、最大值)和离群点。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 boxplot( mtcars$ mpg) fivenum( mtcars$ mpg) [ 1 ] 10.40 15.35 19.20 22.80 33.90 boxplot( mpg ~ cyl, data = mtcars) boxplot( hp ~ cyl, data = mtcars) boxplot( len ~ dose, data = ToothGrowth) boxplot( len ~ dose: supp, data = ToothGrowth) boxplot( mpg ~ cyl, data = mtcars, col = c ( 'pink' , 'orange' , 'red' ) ) boxplot( mpg ~ cyl, data = mtcars, col = c ( 'pink' , 'orange' , 'red' ) , pch = 16 , cex = 0.8 ) boxplot( mpg ~ cyl, data = mtcars, col = c ( 'pink' , 'orange' , 'red' ) , pch = 16 , cex = 0.8 , xlab = 'number of cylinders' , ylab = 'miles/gallon' ) boxplot( len ~ dose, data = ToothGrowth, notch = T ) boxplot( len ~ dose, data = ToothGrowth, width = c ( 0.5 , 1 , 2 ) ) boxplot( len ~ dose, data = ToothGrowth, varwidth = T ) boxplot( len ~ dose, data = ToothGrowth, boxwex = 0.5 ) boxplot( len ~ dose, data = ToothGrowth, staplewex = 0.5 ) boxplot( len ~ dose: supp, data = ToothGrowth, sep = ':' ) boxplot( len ~ dose: supp, data = ToothGrowth, sep = ':' , lex.order = T ) x <- read.csv( '../Rdata/gene_expression.csv' , header = T , row.names = 1 ) head( x) A1 A2 A3 B1 B2 B3 gene1 0.33 0.13 0.30 0.48 0.37 1.73 gene2 0.00 0.00 0.30 0.54 0.43 0.35 gene3 10.27 10.05 5.51 30.95 15.85 25.72 gene4 3.74 4.44 4.24 0.24 0.06 1.04 gene5 9.66 10.05 15.57 6.83 2.66 5.89 gene6 0.50 0.64 0.18 0.72 1.24 2.60 boxplot( x) boxplot( x, outline = F ) library( reshape2) x <- melt( x) No id variables; using all as measure variables head( x) variable value 1 A1 0.33 2 A1 0.00 3 A1 10.27 4 A1 3.74 5 A1 9.66 6 A1 0.50 boxplot( value ~ variable, data = x, outline = F )
热图
生信分析中常见的图,根据颜色的深浅反应数值的大小,可用于基因表达的可视化和聚类。
绘制热图的对象是数值矩阵。
绘制热图的方法:
heatmap()gplot包heatmap.2()pheatmap包pheatmap()ComplexHeatmap包
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 class ( mtcars) [ 1 ] "data.frame" heatmap( mtcars) Error in heatmap( mtcars) : 'x' must be a numeric matrix heatmap( as.matrix( mtcars) ) heatmap( as.matrix( mtcars) , scale = 'col' ) m <- read.csv( '../Rdata/heatmap.csv' , row.names = 1 ) x <- as.matrix( m) heatmap( x) heatmap( t( x) ) heatmap( x, col= c ( 'green' , 'blue' ) ) colorRampPalette( c ( 'red' , 'black' , 'green' ) ) ( nrow( x) ) [ 1 ] "#FF0000" "#ED0000" "#DB0000" "#CA0000" "#B80000" "#A70000" "#950000" "#830000" "#720000" [ 10 ] "#600000" "#4F0000" "#3D0000" "#2B0000" "#1A0000" "#080000" "#000800" "#001A00" "#002B00" [ 19 ] "#003D00" "#004F00" "#006000" "#007200" "#008300" "#009500" "#00A700" "#00B800" "#00CA00" [ 28 ] "#00DB00" "#00ED00" "#00FF00" yanse <- colorRampPalette( c ( 'red' , 'black' , 'green' ) ) ( nrow( x) ) heatmap( x, col = yanse) heatmap( x, col = yanse, ColSideColors = colorRampPalette( c ( 'red' , 'black' , 'green' ) ) ( ncol( x) ) , RowSideColors = yanse) heatmap( x, col = yanse, Colv = NA , Rowv = NA ) library( gplots) m <- as.matrix( read.csv( '../Rdata/heatmap.csv' , row.names = 1 ) ) heatmap.2( m) heatmap.2( m, key = F ) heatmap.2( m, symkey = T ) heatmap.2( m, symkey = F ) heatmap.2( m, symkey = F , density.info = 'none' ) heatmap.2( m, symkey = F , trace = 'none' ) heatmap.2( m, symkey = F , tracecol = 'green' ) heatmap.2( m, dendrogram = 'none' ) heatmap.2( m, dendrogram = 'row' ) heatmap.2( m, dendrogram = 'col' ) library( pheatmap) pheatmap( m) pheatmap( m, border_color = 'white' ) rep ( c ( 'N1' , 'T1' , 'N2' , 'T2' ) , each= 5 ) [ 1 ] "N1" "N1" "N1" "N1" "N1" "T1" "T1" "T1" "T1" "T1" "N2" "N2" "N2" "N2" "N2" "T2" "T2" "T2" [ 19 ] "T2" "T2" factor( rep ( c ( 'N1' , 'T1' , 'N2' , 'T2' ) , each= 5 ) ) [ 1 ] N1 N1 N1 N1 N1 T1 T1 T1 T1 T1 N2 N2 N2 N2 N2 T2 T2 T2 T2 T2 Levels: N1 N2 T1 T2 annotation_col <- data.frame( factor( rep ( c ( 'N1' , 'T1' , 'N2' , 'T2' ) , each= 5 ) ) ) row.names( annotation_col) <- colnames( m) colnames( annotation_col) <- 'Cell Type' head( annotation_col) Cell Type N.GD1 N1 N.GD2 N1 N.GD3 N1 N.GD4 N1 N.GD5 N1 T.GD1 T1
图表设置
图表参数
par函数,Parameter简写,设置绘图中的参数。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 head( par( ) ) $ xlog[ 1 ] FALSE $ ylog[ 1 ] FALSE $ adj[ 1 ] 0.5 $ ann[ 1 ] TRUE $ ask[ 1 ] FALSE $ bg[ 1 ] "white" ? par( ) par( pch= 16 ) opar <- par( no.readonly = T ) par( opar) plot( women$ height, women$ weight, type = "b" , pch= 16 , lty= 2 , lwd= 1.2 , las= 1 , main = "This is main title" , sub = "This is sub title" , xlab = "x label" , ylab = "y label" , xlim = c ( 50 , 100 ) , ylim = c ( 100 , 200 ) , adj= 0.5 , bty= "]" , tck= - 0.03 , col= "red" , fg= "blue" , col.main= "green" , col.sub= 'pink' , col.lab= 'orange' , col.axis= 'purple' , cex= 1.5 , cex.lab= 1.2 , cex.axis= 1.2 , cex.main= 1.5 )
图表颜色
颜色选择标准:
对比明显
低调内敛
色盲友好
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 colors( ) [ 1 ] "white" "aliceblue" "antiquewhite" [ 4 ] "antiquewhite1" "antiquewhite2" "antiquewhite3" [ 7 ] "antiquewhite4" "aquamarine" "aquamarine1" ... sample( colors( ) , 5 ) [ 1 ] "cadetblue4" "palegreen4" "hotpink2" "lightsteelblue2" "gray12" rgb( 0.1 , 0.2 , 0.3 ) [ 1 ] "#1A334D" hsv( 0.1 , 0.2 , 0.3 ) [ 1 ] "#4D463D" hcl( 0.1 , 0.2 , 0.3 ) [ 1 ] "#020101" rainbow( 5 ) [ 1 ] "#FF0000" "#CCFF00" "#00FF66" "#0066FF" "#CC00FF" heat.colors( 5 ) [ 1 ] "#FF0000" "#FF5500" "#FFAA00" "#FFFF00" "#FFFF80" terrain.colors( 5 ) [ 1 ] "#00A600" "#E6E600" "#EAB64E" "#EEB99F" "#F2F2F2" topo.colors( 5 ) [ 1 ] "#4C00FF" "#004CFF" "#00E5FF" "#00FF4D" "#FFFF00" cm.colors( 5 ) [ 1 ] "#80FFFF" "#BFFFFF" "#FFFFFF" "#FFBFFF" "#FF80FF" grey.colors( 5 ) [ 1 ] "#4D4D4D" "#888888" "#AEAEAE" "#CCCCCC" "#E6E6E6" library( RColorBrewer) display.brewer.all( ) display.brewer.pal( name = 'Set3' , n = 5 ) brewer.pal( name = 'Set3' , n = 5 ) [ 1 ] "#8DD3C7" "#FFFFB3" "#BEBADA" "#FB8072" "#80B1D3" pie( 1 : 5 , col = brewer.pal( 'Set3' , n = 5 ) )
韦恩图
一般最少2组,最多5组,再多图很复杂没有意义。
韦恩图画法:
gplots包venn函数(简单,无颜色)venneuler包中的venneuler函数(安装失败)venn包中的venn函数(简单,有颜色,推荐)vennDiagram包中的venn.Diagram函数(有颜色,图没有上个好看,必须输出文件,无法在plots窗口预览)
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 listA <- read.csv( '../Rdata/genes_list_A.txt' , header = F ) listB <- read.csv( '../Rdata/genes_list_B.txt' , header = F ) listC <- read.csv( '../Rdata/genes_list_C.txt' , header = F ) listD <- read.csv( '../Rdata/genes_list_D.txt' , header = F ) listE <- read.csv( '../Rdata/genes_list_E.txt' , header = F ) A <- listA$ V1 B <- listB$ V1 C <- listC$ V1 D <- listD$ V1 E <- listE$ V1 length ( A) ;length ( B) ;length ( C) ;length ( D) ;length ( E) [ 1 ] 562 [ 1 ] 474 [ 1 ] 2273 [ 1 ] 1735 [ 1 ] 594 library( gplots) venn( list ( A, B, C, D, E) ) library( VennDiagram) venn.diagram( list ( sampleC= C, sampleD= D) , filename = 'venn.png' , fill = c ( 'yellow' , 'cyan' ) , cex = 1.5 ) [ 1 ] 1 venn.diagram( list ( sampleC= C, sampleD= D) , filename = 'venn2.png' , fill = c ( 'yellow' , 'cyan' ) , cex = 1.2 , disable.logging = T ) INFO [ 2022 - 02 - 01 17 : 06 : 37 ] [[ 1 ] ] INFO [ 2022 - 02 - 01 17 : 06 : 37 ] list ( sampleC = C, sampleD = D) INFO [ 2022 - 02 - 01 17 : 06 : 37 ] INFO [ 2022 - 02 - 01 17 : 06 : 37 ] $ filename INFO [ 2022 - 02 - 01 17 : 06 : 37 ] [ 1 ] "venn2.png" INFO [ 2022 - 02 - 01 17 : 06 : 37 ] INFO [ 2022 - 02 - 01 17 : 06 : 37 ] $ fill INFO [ 2022 - 02 - 01 17 : 06 : 37 ] c ( "yellow" , "cyan" ) INFO [ 2022 - 02 - 01 17 : 06 : 37 ] INFO [ 2022 - 02 - 01 17 : 06 : 37 ] $ cex INFO [ 2022 - 02 - 01 17 : 06 : 37 ] [ 1 ] 1.2 INFO [ 2022 - 02 - 01 17 : 06 : 37 ] INFO [ 2022 - 02 - 01 17 : 06 : 37 ] $ disable.logging INFO [ 2022 - 02 - 01 17 : 06 : 37 ] T INFO [ 2022 - 02 - 01 17 : 06 : 37 ] [ 1 ] 1 venn.diagram( list ( sampleC= C, sampleD= D, sampleA= A) , filename = 'venn3.png' , fill = c ( 'yellow' , 'cyan' , 'blue' ) , cex = 1.2 , disable.logging = T ) venn.diagram( list ( sampleC= C, sampleD= D, sampleA= A, sampleB= B) , filename = 'venn4.png' , fill = c ( 'yellow' , 'cyan' , 'blue' , 'red' ) , cex = 1.2 , disable.logging = T ) venn.diagram( list ( sampleC= C, sampleD= D, sampleA= A, sampleB= B, sampleE= E) , filename = 'venn5.png' , fill = c ( 'yellow' , 'cyan' , 'blue' , 'red' , 'pink' ) , cex = 1.2 , disable.logging = T ) venn( list ( A, B, C, D, E) ) venn( list ( A, B, C, D, E) , box = F ) venn( list ( A, B, C, D, E) , box = F , col= 'red' ) venn( list ( A, B, C, D, E) , box = F , snames = 'Sample A,Sample B,Sample C,Sample D,Sample E' ) venn( list ( A, B, C, D, E) , box = F , snames = 'Sample A,Sample B,Sample C,Sample D,Sample E' , zcolor = 'red, green, purple, blue, cyan' ) venn( list ( A, B, C, D, E) , box = F , snames = 'Sample A,Sample B,Sample C,Sample D,Sample E' , zcolor = 'red, green, purple, blue, cyan' , ellipse = T )
曼哈顿图
曼哈顿图是用二维散点图展示大量数据的方法,主要用于全基因组关联分析。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 library( qqman) library( RColorBrewer) gwasResults SNP CHR BP P 1 rs1 1 1 0.9148060435 2 rs2 1 2 0.9370754133 3 rs3 1 3 0.2861395348 ... manhattan( gwasResults) manhattan( gwasResults, main = 'Manhattan Plot' , ylim = c ( 0 , 6 ) , cex = 0.6 , cex.axis = 0.9 ) manhattan( gwasResults, main = 'Manhattan Plot' , ylim = c ( 0 , 6 ) , cex = 0.6 , suggestiveline = FALSE , genomewideline = FALSE ) manhattan( gwasResults, main = 'Manhattan Plot' , ylim = c ( 0 , 6 ) , cex = 0.6 , cex.axis= 0.9 , col = c ( 'blue4' , 'orange3' ) ) yanse <- brewer.pal( 'Set3' , n= 4 ) manhattan( gwasResults, main = 'Manhattan Plot' , ylim = c ( 0 , 6 ) , cex = 0.6 , cex.axis= 0.9 , col = yanse, chrlabs = c ( 1 : 20 , 'P' , 'Q' ) ) manhattan( gwasResults, main = 'Manhattan Plot' , ylim = c ( 0 , 6 ) , cex = 0.6 , cex.axis= 0.9 , col = c ( 'blue4' , 'orange3' ) , chrlabs = c ( 1 : 20 , 'P' , 'Q' ) ) manhattan( subset( gwasResults, CHR== 3 ) ) snpsOfInterest [ 1 ] "rs3001" "rs3002" "rs3003" "rs3004" "rs3005" "rs3006" "rs3007" "rs3008" "rs3009" "rs3010" [ 11 ] "rs3011" "rs3012" "rs3013" "rs3014" "rs3015" "rs3016" "rs3017" "rs3018" "rs3019" "rs3020" [ 21 ] "rs3021" "rs3022" "rs3023" "rs3024" "rs3025" "rs3026" "rs3027" "rs3028" "rs3029" "rs3030" [ 31 ] "rs3031" "rs3032" "rs3033" "rs3034" "rs3035" "rs3036" "rs3037" "rs3038" "rs3039" "rs3040" [ 41 ] "rs3041" "rs3042" "rs3043" "rs3044" "rs3045" "rs3046" "rs3047" "rs3048" "rs3049" "rs3050" [ 51 ] "rs3051" "rs3052" "rs3053" "rs3054" "rs3055" "rs3056" "rs3057" "rs3058" "rs3059" "rs3060" [ 61 ] "rs3061" "rs3062" "rs3063" "rs3064" "rs3065" "rs3066" "rs3067" "rs3068" "rs3069" "rs3070" [ 71 ] "rs3071" "rs3072" "rs3073" "rs3074" "rs3075" "rs3076" "rs3077" "rs3078" "rs3079" "rs3080" [ 81 ] "rs3081" "rs3082" "rs3083" "rs3084" "rs3085" "rs3086" "rs3087" "rs3088" "rs3089" "rs3090" [ 91 ] "rs3091" "rs3092" "rs3093" "rs3094" "rs3095" "rs3096" "rs3097" "rs3098" "rs3099" "rs3100" manhattan( gwasResults, highlight = snpsOfInterest) manhattan( gwasResults, annotatePval = 0.001 ) manhattan( gwasResults, annotatePval = 0.001 , annotateTop = FALSE )
火山图
散点图的一种,主要用于基因表达的分析中。
自带散点图
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 m <- read.csv( '../Rdata/Deseq2.csv' , header = T , row.names = 1 ) head( m) baseMean log2FoldChange lfcSE stat pvalue padj ENSG00000000003 708.6021697 - 0.38125397 0.10065597 - 3.7876937 1.520521e-04 0.0012815112 ENSG00000000419 520.2979006 0.20681259 0.11222180 1.8428915 6.534485e-02 0.1962308610 ENSG00000000457 237.1630368 0.03792034 0.14345322 0.2643394 7.915184e-01 0.9112208706 ENSG00000000460 57.9326331 - 0.08816367 0.28716771 - 0.3070111 7.588349e-01 0.8946714454 ENSG00000000938 0.3180984 - 1.37822703 3.49987280 - 0.3937935 6.937335e-01 NA ENSG00000000971 5817.3528677 0.42640216 0.08831006 4.8284666 1.375884e-06 0.0000181641 m <- na.omit( m) plot( m$ log2FoldChange, m$ padj) plot( m$ log2FoldChange, - log10( m$ padj) , pch= 16 , cex= 0.6 , xlim = c ( - 10 , 10 ) , ylim = c ( 0 , 100 ) ) transform( m, padj= - log10( m$ padj) ) baseMean log2FoldChange lfcSE stat pvalue padj ENSG00000000003 708.602170 - 0.3812539747 0.10065597 - 3.787693677 1.520521e-04 2.892278e+00 ENSG00000000419 520.297901 0.2068125935 0.11222180 1.842891491 6.534485e-02 7.072327e-01 ENSG00000000457 237.163037 0.0379203397 0.14345322 0.264339418 7.915184e-01 4.037634e-02 ... m <- transform( m, padj= - log10( m$ padj) ) down <- m[ m$ log2FoldChange <= - 1 , ] up <- m[ m$ log2FoldChange >= 1 , ] mid <- m[ m$ log2FoldChange - 1 & m$ log2FoldChange < 1 , ] plot( mid$ log2FoldChange, mid$ padj, xlim = c ( - 10 , 10 ) , ylim = c ( 0 , 100 ) , pch = 16 , cex = 0.6 , col = 'grey' , main = 'Gene Expression' , xlab = 'log2FodeChange' , ylab = '-log10(Qvalue)' ) points( up$ log2FoldChange, up$ padj, pch= 16 , cex= 0.6 , col= 'red' ) points( down$ log2FoldChange, down$ padj, pch= 16 , cex= 0.6 , col= 'green' )
ggplot包
1 2 3 4 5 m <- read.csv( '../Rdata/Deseq2.csv' , header = T , row.names = 1 ) m <- na.omit( m) transform( m, padj= - log10( m$ padj) ) m <- transform( m, padj= - log10( m$ padj) )
ggplot2作图
逻辑清晰,绘图方便,简介好看,有很多基于ggplot2的绘图包。
图形语法:grammar of graphis,类似于图层的概念。
1.数据(data)
一般为数据框
2.映射(mapping)
可以将size,shape,col等映射为数据框中的值
1 2 3 4 ggplot( data = mtcars, mapping = aes( x= wt, y= mpg) ) ggplot( data = mtcars, mapping = aes( x= wt, y= mpg, size= mpg) ) ggplot( data = mtcars, mapping = aes( x= wt, y= mpg, shape= as.factor( cyl) ) ) ggplot( data = mtcars, mapping = aes( x= wt, y= mpg, col= mpg) )
3.几何对象
1 ggplot( data = mtcars, mapping = aes( x= wt, y= mpg) ) + geom_point( )